 |
Description: Argonautes and small RNAs associated with nematode programmed DNA elimination
Author: MVZ
Session Name: small_RNAs
Genome Assembly: hub_4238208_v37
Creation Date: 2025-12-13
Views: 21
|
1765612800 |
21 |
 |
Description: Anti-BATF ChIP on FACS-purified, bone marrow–derived pDCs at steady state and after 2 h of CpG stimulation.
Author: AliSh
Session Name: BATF_pDCs
Genome Assembly: mm10
Creation Date: 2025-12-11
Views: 22
|
1765440000 |
22 |
 |
Description: for final presentation
Author: ashelhorse
Session Name: ClinicalVarSCN9A
Genome Assembly: hg38
Creation Date: 2025-12-09
Views: 66
|
1765267200 |
66 |
 |
Description: Annotation of LCT and MCM6 Genes
Author: Caesaram
Session Name: LCT and MCM6 Gene (F)
Genome Assembly: hg38
Creation Date: 2025-12-09
Views: 23
|
1765267200 |
23 |
 |
Description: HBB gene showing snips where sickle cell mutation occurs, as well as graph showing how HBB is only expressed in blood cells
Author: allygarrett
Session Name: hg38 - HBB gene
Genome Assembly: hg38
Creation Date: 2025-12-04
Views: 25
|
1764835200 |
25 |
 |
Description: CRX uniprot assignment
Author: allygarrett
Session Name: hg38 - CRX protein Uniprot
Genome Assembly: hg38
Creation Date: 2025-12-02
Views: 28
|
1764662400 |
28 |
 |
Description: isoforms figure 1
Author: sronalds
Session Name: CRX_isoforms
Genome Assembly: hg38
Creation Date: 2025-12-02
Views: 23
|
1764662400 |
23 |
 |
Description: View of CRX gene and its isoforms with tracks enabled to show possible mutations
Author: messicbr
Session Name: hg38 CRX w/ splice and isoforms
Genome Assembly: hg38
Creation Date: 2025-12-02
Views: 21
|
1764662400 |
21 |
 |
Description: CRX gene isoforms
Author: MadisonTesnow
Session Name: hg38 CRX gene isoforms Madison Tesnow
Genome Assembly: hg38
Creation Date: 2025-12-02
Views: 8
|
1764662400 |
8 |
 |
Description: pde6c/rbp4
Author: katelynnmenjivar
Session Name: mm9 KM PDE6C/RBP4 locus
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 74
|
1763452800 |
74 |
 |
Description: PDE6C/RBP4 locus mm9
Author: oliviakloc
Session Name: PDE6C/RBP4 locus mm9
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 31
|
1763452800 |
31 |
 |
Description: guca1b/guca1a
Author: katelynnmenjivar
Session Name: mm9 KM GUCA1B/GUCA1A locus
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 27
|
1763452800 |
27 |
 |
Description: nrl/cpne6
Author: katelynnmenjivar
Session Name: mm9 KM NRL/CPNE6 locus
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 11
|
1763452800 |
11 |
 |
Description: rho/h1foo
Author: katelynnmenjivar
Session Name: mm9 KM RHO/H1FOO
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 14
|
1763452800 |
14 |
 |
Description: In class assignment view of RHO/H1FOO with CBRs, mRNAs, and DHS.
Author: LucaDooley
Session Name: mm9 RHO/H1FOO CBR,mRNA&DHS
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: Had to delete the other this is the working view
Author: sronalds
Session Name: PDE6C/RBP4_SRonalds_BIO581_Real
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: In class assignment view of PDE6C/RBP4 with CBRs, mRNAs, and DHS.
Author: LucaDooley
Session Name: mm9 PDE6C/RBP4 CBR,mRNA&DHS
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 12
|
1763452800 |
12 |
 |
Description: View of the fully RHO gene with custom tracks for wild type and NRL enabled
Author: messicbr
Session Name: mm9 RHO_WT/NRL
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 12
|
1763452800 |
12 |
 |
Description: mm9 CRX, mRNA, Dnase1 on NRL
Author: Elizabeth McLaughlin
Session Name: mm9 CRX, mRNA, Dnase1 on NRL
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: Good zooming
Author: sronalds
Session Name: NRL/CPNE6_Sronalds_BIO581
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 8
|
1763452800 |
8 |
 |
Description: PDE6C
Author: Caesaram
Session Name: CBR/mRNA PDE6C day1week1week8
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 5
|
1763452800 |
5 |
 |
Description: NRL
Author: Caesaram
Session Name: CBR/mRNA NRL day1week1week8
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: GUCA1B/GUCA1A locus mm9
Author: oliviakloc
Session Name: GUCA1B/GUCA1A locus mm9
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 6
|
1763452800 |
6 |
 |
Description: Not super zoomed in
Author: sronalds
Session Name: GUCA1B/A_SRonalds_BIO581
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 9
|
1763452800 |
9 |
 |
Description: all data
Author: katelynnmenjivar
Session Name: mm9 all data KM
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 12
|
1763452800 |
12 |
 |
Description: In class assignment view of NRL/CPNE6 with CBRs, mRNAs, and DHS.
Author: LucaDooley
Session Name: mm9 NRL/CPNE6 CBR,mRNA&DHS
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: In class assignment view of GUCA1a/1b with CBRs, mRNAs, and DHS.
Author: LucaDooley
Session Name: mm9 GUCA1a/1b CBR,mRNA&DHS
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: NRL/CPNE6 locus mm9
Author: oliviakloc
Session Name: NRL/CPNE6 locus mm9
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 11
|
1763452800 |
11 |
 |
Description: super zoomed in
Author: sronalds
Session Name: RHO/H1FOO_Sronalds_BIO581
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 9
|
1763452800 |
9 |
 |
Description: GUCA
Author: Caesaram
Session Name: CBR/mRNA GUCA day1week1week8
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: Fixed view of the NRL/CPNE6 genes with custom tracks for wild type and NRL retina tissue as well as CBR tracks
Author: messicbr
Session Name: mm9 NRL/CPNE6 w/ NR1 (updated)
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: CBR and mRNA in class practice
Author: allygarrett
Session Name: mm9 CBR and mRNA - AGarrett
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: mm9 everything PDE6C
Author: Elizabeth Callis
Session Name: mm9 everything PDE6C
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 6
|
1763452800 |
6 |
 |
Description: mm9 everything GUCA1B
Author: Elizabeth Callis
Session Name: mm9 everything GUCA1B
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: DHS seq mm9 on RHO gene
Author: oliviakloc
Session Name: Combined tracks- DHS seq mm9
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: mm9 everything NRL
Author: Elizabeth Callis
Session Name: mm9 everything NRL
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: CBR practice
Author: ashelhorse
Session Name: mm9.3
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 8
|
1763452800 |
8 |
 |
Description: in class assignment 11/18
Author: lgunter15
Session Name: mm9_mRNA_custom_tracks_gunter
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: View of PDE6c Exons 1-16 showing custom wt and NR1 retina tracks with complementary CBR tracks.
Author: messicbr
Session Name: mm9 PDE6C
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: mm9 everything RHO
Author: Elizabeth Callis
Session Name: mm9 everything RHO
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 11
|
1763452800 |
11 |
 |
Description: The template that will be used for the figures for 11/18/2025 class (have chip seq and rna seq custom tracks)
Author: sronalds
Session Name: RHO_CRX_Fig1_Template_SRonalds_BIO581
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 11
|
1763452800 |
11 |
 |
Description: wt & Nrl-
Author: katelynnmenjivar
Session Name: mm9 mouse wt & Nrl-
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: View of GUCA1B and GUCA1A with custom tracks for wt retina and NR1 retina, with DNase 1 hypersensitivity enabled
Author: messicbr
Session Name: mm9 GUCA1B/GUCA1B
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 9
|
1763452800 |
9 |
 |
Description: All custom tracks with Nrl- RNA-seq, wt RNA-seq, and ChIP-seq data
Author: oliviakloc
Session Name: CRX CHIP-seq mm9
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 8
|
1763452800 |
8 |
 |
Description: View of NRL and CPNE6 with custom track for wt and NR1 retinas
Author: messicbr
Session Name: mm9 NRL/CPNE6 w/ NR1
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 12
|
1763452800 |
12 |
 |
Description: Combined CBR and mRNA data
Author: LucaDooley
Session Name: mm9 RHO CBR&mRNA
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 15
|
1763452800 |
15 |
 |
Description: View of MM9 Exon 1 of RHO with custom tracks showing NR1 Retina
Author: messicbr
Session Name: mm9_RHO Exon1 NR1
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: d1 expression, wk1 expression, wk2 expression
Author: Caesaram
Session Name: CBR/mRNA RHO day1week1week8
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 13
|
1763452800 |
13 |
 |
Description: mm9 wt and nrl CRX binding sites
Author: MadisonTesnow
Session Name: mm9 wt and nrl CRX binding sites Madison Tesnow
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 8
|
1763452800 |
8 |
 |
Description: mm9 Chip-seq + mRNA
Author: Elizabeth Callis
Session Name: mm9 Chip-seq + mRNA
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 8
|
1763452800 |
8 |
 |
Description: CRX binding regions and mRNA expression in the RHO gene
Author: Caesaram
Session Name: CBR/mRNA RHO Mouse mm9
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 8
|
1763452800 |
8 |
 |
Description: mouse genome in-class assignment
Author: lgunter15
Session Name: mm9_Rho_mouse_retina_gunter
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 6
|
1763452800 |
6 |
 |
Description: View of wt and Nrl- CRX Binding sites on RHO.
Author: LucaDooley
Session Name: mm9 RHO CBRs
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 7
|
1763452800 |
7 |
 |
Description: In class
Author: sronalds
Session Name: RHO_CRX_Binding_SRonalds_BIO581
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: aw in class crb mm9
Author: averywitt
Session Name: mm9 CRB
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 8
|
1763452800 |
8 |
 |
Description: CBRs in wildtype and Nrl-
Author: oliviakloc
Session Name: CBRs mm9
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 8
|
1763452800 |
8 |
 |
Description: mm9 CRX activity
Author: allygarrett
Session Name: mm9 CBR - Allison Garrett
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: mm9 RHO 2
Author: Elizabeth Callis
Session Name: mm9 RHO 2
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 10
|
1763452800 |
10 |
 |
Description: CRX binding regions along RHO gen in Mouse
Author: Caesaram
Session Name: CBR RHO Mouse mm9
Genome Assembly: mm9
Creation Date: 2025-11-18
Views: 12
|
1763452800 |
12 |
 |
Description: PDE6B
Author: katelynnmenjivar
Session Name: hg38 PDE6B-KM
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 11
|
1763020800 |
11 |
 |
Description: PDE6A
Author: katelynnmenjivar
Session Name: hg38 PDE6A-KM
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 19
|
1763020800 |
19 |
 |
Description: GNAT2
Author: katelynnmenjivar
Session Name: hg38 GNAT 2-KM
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 9
|
1763020800 |
9 |
 |
Description: PDE6B rod and cone mRNA
Author: oliviakloc
Session Name: PDE6B exons 1-3 hg38
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 15
|
1763020800 |
15 |
 |
Description: GNAT1
Author: katelynnmenjivar
Session Name: hg38 GNAT1-KM
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 14
|
1763020800 |
14 |
 |
Description: dsdawd
Author: sronalds
Session Name: PDE6A_BIO581_SRonalds
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 12
|
1763020800 |
12 |
 |
Description: hg38 PDE6B gene
Author: Elizabeth Callis
Session Name: hg38 PDE6B gene
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 10
|
1763020800 |
10 |
 |
Description: Bam plot for macular and peripheral retina on for GNAT 2 expression.
Author: LucaDooley
Session Name: hg38 bam plot GNAT2
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 20
|
1763020800 |
20 |
 |
Description: hg38 GNAT2
Author: Elizabeth McLaughlin
Session Name: hg38 GNAT2
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 22
|
1763020800 |
22 |
 |
Description: PDE6A rod and cone mRNA
Author: oliviakloc
Session Name: PDE6A exon2 hg38
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 9
|
1763020800 |
9 |
 |
Description: hg38 GNAT1 exon 1
Author: Elizabeth McLaughlin
Session Name: hg38 GNAT1 exon 1
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 16
|
1763020800 |
16 |
 |
Description: View of Exons 20-22 of PDE6B with custom macula and retinal tracks enabled
Author: messicbr
Session Name: hg38 PDE6B Exons 20-22 BM
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 11
|
1763020800 |
11 |
 |
Description: hg38 GNAT 2 gene
Author: Elizabeth Callis
Session Name: hg38 GNAT 2 gene
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 11
|
1763020800 |
11 |
 |
Description: hg38 PDE6A exon 2
Author: Elizabeth McLaughlin
Session Name: hg38 PDE6A exon 2
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 22
|
1763020800 |
22 |
 |
Description: hg38 GNAT 1 gene
Author: Elizabeth Callis
Session Name: hg38 GNAT 1 gene
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 12
|
1763020800 |
12 |
 |
Description: PED6a 1st 3 exons, macular and peripheral degeneration
Author: Will42ka
Session Name: PED6A_1st3_exons
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 11
|
1763020800 |
11 |
 |
Description: hg38 PDE6E exons 1-3
Author: Elizabeth McLaughlin
Session Name: hg38 PDE6B exons 1-3
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 19
|
1763020800 |
19 |
 |
Description: View of GNAT2 with macular and retinal custom tracks enabled
Author: messicbr
Session Name: hg38_GNAT2 BM
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 12
|
1763020800 |
12 |
 |
Description: GNAT1 rod and cone mRNA
Author: oliviakloc
Session Name: GNAT1 exon1 hg38
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 12
|
1763020800 |
12 |
 |
Description: hhh
Author: sronalds
Session Name: GNAT1_BIO581_Sronalds
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 15
|
1763020800 |
15 |
 |
Description: View of GNAT1 Exon 1 with Macular and Retinal custom tracks
Author: messicbr
Session Name: hg38_GNAT1_Exon1 Messick
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 13
|
1763020800 |
13 |
 |
Description: GNAT2 rod and cone mRNA
Author: oliviakloc
Session Name: GNAT2 hg38
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 9
|
1763020800 |
9 |
 |
Description: gnat1 1st exon, macular and peripheral degeneration
Author: Will42ka
Session Name: hg38_gnat1_1st_exon
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 13
|
1763020800 |
13 |
 |
Description: View of the second exon of PDE6A with Retinal and Macular Custom Tracks enabled
Author: messicbr
Session Name: hg38 PDE6A_Exon2 BM
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 14
|
1763020800 |
14 |
 |
Description: hg38 PDE6A gene
Author: Elizabeth Callis
Session Name: hg38 PDE6A gene
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 17
|
1763020800 |
17 |
 |
Description: Custom Tracks
Author: Caesaram
Session Name: Mac/Ret Gnat2
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 13
|
1763020800 |
13 |
 |
Description: Custom Tracks
Author: Caesaram
Session Name: Mac/Ret Gnat1
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 12
|
1763020800 |
12 |
 |
Description: Custom Tracks
Author: Caesaram
Session Name: Mac/Ret PDE6A
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 14
|
1763020800 |
14 |
 |
Description: gnat 2, macular and peripheral degeneration
Author: Will42ka
Session Name: hg38_Gnat2_mac_per
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 13
|
1763020800 |
13 |
 |
Description: Custom Tracks
Author: Caesaram
Session Name: Mac/Ret PDE6B
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 12
|
1763020800 |
12 |
 |
Description: Practice BAM files using cones and rods
Author: allygarrett
Session Name: Practice Bam - Allison Garrett
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 9
|
1763020800 |
9 |
 |
Description: PED6b 1st 3 exons, macular and peripheral degeneration
Author: Will42ka
Session Name: hg38_pde6b_1st3_exons
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 17
|
1763020800 |
17 |
 |
Description: blhgfhyj
Author: sronalds
Session Name: GNAT2_BIO581_SRonalds
Genome Assembly: hg38
Creation Date: 2025-11-13
Views: 13
|
1763020800 |
13 |
 |
Description: hg38 MAPT gene custom track Gencode v49
Author: MadisonTesnow
Session Name: FINAL hg38 MAPT gene custom track Gencode v49
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 37
|
1762848000 |
37 |
 |
Description: MAPT mutant alleles found on chromosome 17 in the human hg38 assembly
Author: ashelhorse
Session Name: MAPTcustumtrackAS
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 21
|
1762848000 |
21 |
 |
Description: Practice for making and editing custom tracks
Author: allygarrett
Session Name: hg38 MAPT custom tracks
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 29
|
1762848000 |
29 |
 |
Description: BED data exercise
Author: ashelhorse
Session Name: hg38 2013 MAPT mutationsAS
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 14
|
1762848000 |
14 |
 |
Description: in class custom track
Author: averywitt
Session Name: hg38 MAPT custom track AW
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 10
|
1762848000 |
10 |
 |
Description: in class assignment 11/11
Author: lgunter15
Session Name: hg38_MAPT_assignment_gunter
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 19
|
1762848000 |
19 |
 |
Description: hg38
Author: sronalds
Session Name: Fall25_CustomTrackhg38_Group6_BIO581
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 12
|
1762848000 |
12 |
 |
Description: MAPT mutations custom track
Author: oliviakloc
Session Name: Custom track MAPT hg38
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 10
|
1762848000 |
10 |
 |
Description: custom track, mutations in mapt
Author: Will42ka
Session Name: hg38_custom_track
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 10
|
1762848000 |
10 |
 |
Description: MAPT gene with mutant alleles
Author: Elizabeth Callis
Session Name: MAPT Gene
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 20
|
1762848000 |
20 |
 |
Description: mapt
Author: alexandratrias
Session Name: hg38_MAPT
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 13
|
1762848000 |
13 |
 |
Description: custom data
Author: zshiff
Session Name: hg38-customdata-11/11
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 17
|
1762848000 |
17 |
 |
Description: MAPT Custom Track with GENECODE V36 w/o splice variants enabled.
Author: messicbr
Session Name: BM MAPT Custom Track
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 7
|
1762848000 |
7 |
 |
Description: hg38 MAPT mutation alleles custom track on 11/11
Author: Elizabeth McLaughlin
Session Name: hg38 MAPT mutation alleles custom track 11/11
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 11
|
1762848000 |
11 |
 |
Description: in class custom track
Author: averywitt
Session Name: hg19 RHO custom AW
Genome Assembly: hg19
Creation Date: 2025-11-11
Views: 13
|
1762848000 |
13 |
 |
Description: hg19
Author: sronalds
Session Name: Fall25_CustomTrackhg19_Group6_BIO581
Genome Assembly: hg19
Creation Date: 2025-11-11
Views: 9
|
1762848000 |
9 |
 |
Description: hg19 Human RHO gene Custom track
Author: MadisonTesnow
Session Name: hg19 Human RHO gene Custom track
Genome Assembly: hg19
Creation Date: 2025-11-11
Views: 10
|
1762848000 |
10 |
 |
Description: hg38 MAPT mutant alleles 11/11/25
Author: Delleeb
Session Name: hg38 MAPT Mutations 11/11/25
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 9
|
1762848000 |
9 |
 |
Description: Hg 38 chr17 custome track practice
Author: Caesaram
Session Name: hg38 custom track
Genome Assembly: hg38
Creation Date: 2025-11-11
Views: 10
|
1762848000 |
10 |
 |
Description: class assignment
Author: shifflzk
Session Name: mm9 11/6
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 13
|
1762416000 |
13 |
 |
Description: Trpm1 isoforms in mouse genome
Author: oliviakloc
Session Name: Trpm1 mm9
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 8
|
1762416000 |
8 |
 |
Description: Figure 2
Author: allygarrett
Session Name: mm9 Figure 2 AGarrett
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 10
|
1762416000 |
10 |
 |
Description: rho Trpm1
Author: ashelhorse
Session Name: mm9.2
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 9
|
1762416000 |
9 |
 |
Description: Trpm1 gene mouse
Author: MadisonTesnow
Session Name: Trpm1 gene mouse figure 2 madison tesnow
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 17
|
1762416000 |
17 |
 |
Description: in class figure 2
Author: averywitt
Session Name: mm9 trpm1 aw
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 14
|
1762416000 |
14 |
 |
Description: mouse mm9 Trpm 1 class assignment
Author: katelynnmenjivar
Session Name: mm9 Trpm1-KM
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 14
|
1762416000 |
14 |
 |
Description: rho mm9
Author: ashelhorse
Session Name: mm9
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 19
|
1762416000 |
19 |
 |
Description: trpm1
Author: sronalds
Session Name: Fall25_trpm1_BIO581_Group6
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 7
|
1762416000 |
7 |
 |
Description: mouse mm9 rho in class assignment
Author: katelynnmenjivar
Session Name: mm9 rho-KM
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 11
|
1762416000 |
11 |
 |
Description: trpm1 gene
Author: Will42ka
Session Name: mm9_trpm1_
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 11
|
1762416000 |
11 |
 |
Description: Mouse Trpm 1 gene with day 1, week 1, and week 8 DNase 1 HS
Author: Elizabeth McLaughlin
Session Name: mm9 Trpm1 DNase 1 HS
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 17
|
1762416000 |
17 |
 |
Description: A Garrett mm9 figure 1
Author: allygarrett
Session Name: mm9 Figure 1 AGarrett
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 17
|
1762416000 |
17 |
 |
Description: in class 11/06
Author: lgunter15
Session Name: mm9_gunter_public
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 13
|
1762416000 |
13 |
 |
Description: mm9 Trpm1 viewing DNase1 Hypersensitivity
Author: LucaDooley
Session Name: mm9 Trpm1 DH1
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 13
|
1762416000 |
13 |
 |
Description: rho DNaseI Hs
Author: Will42ka
Session Name: mm9_rho_encode
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 9
|
1762416000 |
9 |
 |
Description: Mouse Trpm1 Gene with UCSC Genes track w/ splice variants enabled and zoomed out 1.5x
Author: messicbr
Session Name: mm9 Trpm1_481BM
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 15
|
1762416000 |
15 |
 |
Description: Mouse Rho gene with day 1, week 1, and week 8 DNase 1 HS
Author: Elizabeth McLaughlin
Session Name: mm9 RHO DNase1 HS
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 12
|
1762416000 |
12 |
 |
Description: mm9 retina mouse gene
Author: MadisonTesnow
Session Name: mm9 retina mouse gene figure 1 Madison Tesnow
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 14
|
1762416000 |
14 |
 |
Description: mm9 Rhodopsin Gene looking at changes from day 1 to 8 weeks for mouse
Author: messicbr
Session Name: mm9 RHO_481BM
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 8
|
1762416000 |
8 |
 |
Description: mouse RHO gene
Author: oliviakloc
Session Name: mm9RHO
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 9
|
1762416000 |
9 |
 |
Description: mess up on discription of last one
Author: Caesaram
Session Name: mm9 rho dnase real
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 13
|
1762416000 |
13 |
 |
Description: in class rho mm9 mouse aw
Author: averywitt
Session Name: mm9 rho aw
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 12
|
1762416000 |
12 |
 |
Description: Mouse rho
Author: sronalds
Session Name: Fall25_MouseMM9_RHO_BIO581_Group6
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 16
|
1762416000 |
16 |
 |
Description: mm9 Rho DNase1H 11/6/25 ED
Author: Delleeb
Session Name: mm9 Rho DNase1H Data 11/6/25
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 19
|
1762416000 |
19 |
 |
Description: mm9 Trpm1 gene
Author: Elizabeth Callis
Session Name: mm9 Trpm1 gene
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 13
|
1762416000 |
13 |
 |
Description: mm9 Trpm1 DNase1 11/6/25 Ed
Author: Delleeb
Session Name: mm9 Trpm1 DNase1H Data 11/6/25
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 24
|
1762416000 |
24 |
 |
Description: mm9 RHO viewing DNaseI Hypersensitivity
Author: LucaDooley
Session Name: mm9 RHO DH1
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 9
|
1762416000 |
9 |
 |
Description: DNase mouse genome
Author: Elizabeth Callis
Session Name: mm9 DNase
Genome Assembly: mm9
Creation Date: 2025-11-06
Views: 13
|
1762416000 |
13 |
 |
Description: Custom Track for Quiz 8 in northeastern 6310 Fall 2025.
Author: fengkailin
Session Name: fk6310q8
Genome Assembly: hg38
Creation Date: 2025-11-03
Views: 88
|
1762156800 |
88 |
 |
Description: CUSTOM TRACKS
Author: mollina
Session Name: hg38_MOLLINA
Genome Assembly: hg38
Creation Date: 2025-11-02
Views: 52
|
1762070400 |
52 |
 |
Description: For Module 08 Quiz:
test1.bed description:
spacer Blue ticks every 10000 bases bed 3 chr22:
even Red ticks every 100 bases, skip 100 bed 3 chr22:
Author: ham.is
Session Name: module08assignment
Genome Assembly: hg38
Creation Date: 2025-11-01
Views: 19
|
1761984000 |
19 |
 |
Description: assignment 8 test.bed annotation of chr 22
Author: vdanthur1
Session Name: quiz8
Genome Assembly: hg38
Creation Date: 2025-10-30
Views: 15
|
1761811200 |
15 |
 |
Description: the genes are left in BED mode
Author: customtracks
Session Name: hg38_bed
Genome Assembly: hg38
Creation Date: 2025-10-29
Views: 34
|
1761724800 |
34 |
 |
Description: bigwig profiles for Zmynd8 study.
Author: gongwx
Session Name: Zmynd8_mm10
Genome Assembly: mm10
Creation Date: 2025-10-29
Views: 65
|
1761724800 |
65 |
 |
Description: Test
Author: marcwayn
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2025-10-27
Views: 124
|
1761552000 |
124 |
 |
Description: a
Author: gregfar
Session Name: CYP2D6_UMAP24_repeats
Genome Assembly: hg38
Creation Date: 2025-10-24
Views: 17
|
1761292800 |
17 |
 |
Description: CSNK2A1_UMAP24_repeats
Author: gregfar
Session Name: CSNK2A1_UMAP24_repeats2
Genome Assembly: hg38
Creation Date: 2025-10-24
Views: 9
|
1761292800 |
9 |
 |
Description: a
Author: gregfar
Session Name: CSNK2A1_UMAP24_repeats
Genome Assembly: hg38
Creation Date: 2025-10-24
Views: 8
|
1761292800 |
8 |
 |
Description: CSNK2A1
Author: gregfar
Session Name: CSNK2A1_UMAP24
Genome Assembly: hg38
Creation Date: 2025-10-22
Views: 112
|
1761120000 |
112 |
 |
Description: bloublou
Author: Michelp
Session Name: Marlene_01
Genome Assembly: sacCer3
Creation Date: 2025-10-20
Views: 47
|
1760947200 |
47 |
 |
Description: A study of the bizarre naked mole rat shows that the animals have evolved a DNA repair mechanism that could explain their longevity. Naked mole-rats can live for up to 30-40 years, which is roughly ten times longer than typical rats, which live only 1–3 years.
When a cell senses damage, one of the substances it produces is a protein called c-GAS. Unlike human cGAS, which inhibits DNA repair by homologous recombination in the nucleus, the naked mole rat enzyme has four amino acid changes that allow it to enhance DNA repair, thus delaying aging and enhancing lifespan, the study concludes.
The four specific amino acid residue substitutions are located within the C-terminal domain of the cGAS protein. This amino acid alteration enabled naked mole-rat cGAS to prolong its retention on chromatin in the wake of DNA damage by modulating its ubiquitination status -which usually signals a protein is ready to be degraded. The changes altered its interaction with segregase P97, which breaks down unneeded or damaged proteins through proteolysis, allowing cGAS to participate in further DNA repair mechanisms. Experimentally reverting these four amino acid residues abolished these protective effects in naked mole rats.
The four amino acids in conferring prolonged chromatin binding (E, Glu; S, Ser; T, Thr; Y, Tyr) and their numbered location on the cGAS protein are S463, E511, Y527, T530. Phylogenetic analysis of cGAS protein sequences across three major rodent clades revealed that the four amino acid changes evolved gradually within the naked mole-rat, but are not observed in other rodents or long-lived mammalian species outside Rodentia. Two exceptions include the gray squirrel (Sciurus carolinensis) and the blind mole-rat (Spalax galili), which have two of the four mutations.
Supplemental data Fig. S2. C shows the multiple amino acid sequence alignment of cGAS C3 region among NM-R, rat, mouse, and human.
Human mutations for cGAS would be D431S (S463), K479E (E511), L495Y (Y527), K498T (T530). The following HGVS notation can be searched in the UCSC Genome Browser to find these spots:
NP_612450.2(CGAS):p.D431S
NP_612450.2(CGAS):p.K479E
NP_612450.2(CGAS):p.L495Y
NP_612450.2(CGAS):p.K498T
Author: brianlee
Session Name: Naked_Mole_Rat_Mutations
Genome Assembly: hg38
Creation Date: 2025-10-14
Views: 129
|
1760428800 |
129 |
 |
Description: this is figure 1
Author: lgunter15
Session Name: HTT_simple_repeates
Genome Assembly: hg38
Creation Date: 2025-10-09
Views: 29
|
1759996800 |
29 |
 |
Description: this is the HTT gene with a zoomed view of the CAG repeats in the gene that cause huntington's.
Author: lgunter15
Session Name: HTT_repeats
Genome Assembly: hg38
Creation Date: 2025-10-09
Views: 29
|
1759996800 |
29 |
 |
Description: assignment for 10/9 with highlighted region of the repeat sequence
Author: katelynnmenjivar
Session Name: hg38 hw 10/9 KM
Genome Assembly: hg38
Creation Date: 2025-10-09
Views: 9
|
1759996800 |
9 |
 |
Description: Figure 1
Author: allygarrett
Session Name: hg38 - F1
Genome Assembly: hg38
Creation Date: 2025-10-08
Views: 61
|
1759910400 |
61 |
 |
Description: Figure 2
Author: allygarrett
Session Name: hg38 - F2
Genome Assembly: hg38
Creation Date: 2025-10-08
Views: 46
|
1759910400 |
46 |
 |
Description: hg38 HTT gene exon 1
Author: Elizabeth Callis
Session Name: hg38 HTT gene exon 1
Genome Assembly: hg38
Creation Date: 2025-10-08
Views: 17
|
1759910400 |
17 |
 |
Description: HTT in class assignment due 10/9
Author: averywitt
Session Name: HTT in class
Genome Assembly: hg38
Creation Date: 2025-10-08
Views: 14
|
1759910400 |
14 |
 |
Description: Looking for simple repeats on Chr 4 of HTT gene.
Author: SamiraL
Session Name: HTT with “Simple Repeats” figure 1
Genome Assembly: hg38
Creation Date: 2025-10-08
Views: 16
|
1759910400 |
16 |
 |
Description: HTT Gene Zoomed in on CAG repeat region
Author: messicbr
Session Name: hg38 HTT_ZoomedIN
Genome Assembly: hg38
Creation Date: 2025-10-08
Views: 8
|
1759910400 |
8 |
 |
Description: HTT Gene with GENCODE V46 and Simple Repeats
Author: messicbr
Session Name: hg38 HTT_ZoomedOut
Genome Assembly: hg38
Creation Date: 2025-10-08
Views: 10
|
1759910400 |
10 |
 |
Description: Simple Repeats track in squish view shows multiple repeat elements distributed throughout the HTT gene, including CAG repeat regions implicated in Huntington’s disease.
Author: ashelhorse
Session Name: HTTFig1
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 15
|
1759824000 |
15 |
 |
Description: This snapshot displays the CAG repeat region in the first exon of HTT. The Simple Repeats track reports about 21 CAG repeats, and the Short Match tool highlights tandem CAG sequences. Because this number is well below the disease threshold (>39 repeats), the reference genome individual would not be expected to develop Huntington’s disease.
Author: ashelhorse
Session Name: HTTFig2
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 13
|
1759824000 |
13 |
 |
Description: Figure 2
Author: sronalds
Session Name: Fall25_HTTgene_Group6_BIO581_Fig2
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 9
|
1759824000 |
9 |
 |
Description: HTT gene repeat polymorphism associated with Huntington’s disease
Author: ashelhorse
Session Name: HTT
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 28
|
1759824000 |
28 |
 |
Description: HTT gene overview for class
Author: LucaDooley
Session Name: hg38 HTT Gene
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 13
|
1759824000 |
13 |
 |
Description: Human HTT gene annotated
Author: Caesaram
Session Name: hg38 HTT
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 14
|
1759824000 |
14 |
 |
Description: Before class quiz
Author: sronalds
Session Name: Fall25_HTTgene_Quiz_BIO581
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 7
|
1759824000 |
7 |
 |
Description: HTT Gene - Allison Garrett
Author: allygarrett
Session Name: hg38-HTT
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 14
|
1759824000 |
14 |
 |
Description: Figure 1
Author: sronalds
Session Name: Fall25_HTT_Group6_BIO581_Fig1
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 14
|
1759824000 |
14 |
 |
Description: HTT gene with view of simple repeats quiz for 10/07
Author: lgunter15
Session Name: hg38_HTT
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 13
|
1759824000 |
13 |
 |
Description: HTT
Author: shifflzk
Session Name: hg38. HTT
Genome Assembly: hg38
Creation Date: 2025-10-06
Views: 12
|
1759737600 |
12 |
 |
Description: HTT Gene with Genecode V32 and Simple Repeats tracks
Author: messicbr
Session Name: hg38 HTT Gene
Genome Assembly: hg38
Creation Date: 2025-10-06
Views: 14
|
1759737600 |
14 |
 |
Description: 10/03 hw assignment
Author: katelynnmenjivar
Session Name: hg38 hw 10/3
Genome Assembly: hg38
Creation Date: 2025-10-06
Views: 12
|
1759737600 |
12 |
 |
Description: Looking at Huntington’s disease CHR 4.
Author: SamiraL
Session Name: SL Huntington’s disease Hg38/13
Genome Assembly: hg38
Creation Date: 2025-10-06
Views: 11
|
1759737600 |
11 |
 |
Description: HW 10/6 HTT gene
Author: averywitt
Session Name: hg38 HTT gene AW
Genome Assembly: hg38
Creation Date: 2025-10-06
Views: 11
|
1759737600 |
11 |
 |
Description: Displays two custom BED tracks (spacer and even) on chr22:20,100,000–20,100,100.
Blue ticks every 10 kb, red ticks every 100 bp.
Author: Sayedahmed.a
Session Name: hg38-test_bed_assignment
Genome Assembly: hg38
Creation Date: 2025-10-27
Views: 23
|
1761552000 |
23 |
 |
Description: the HTT gene in the human hg38 genome browser.
Author: Elizabeth Callis
Session Name: hg38 HTT gene
Genome Assembly: hg38
Creation Date: 2025-10-03
Views: 18
|
1759478400 |
18 |
 |
Description: This UCSC public session provides interactive visualization of the complete ChIP-seq and bulk ATAC-seq datasets presented in Mariner et al., manuscript submitted to Nucleic Acids Research.
Author: mariamafau
Session Name: Mariner_NAR_2025
Genome Assembly: mm10
Creation Date: 2025-09-23
Views: 53
|
1758614400 |
53 |
 |
Description: primates have similar/different chromosome fusion events to the human-chimp fusion relationship
Author: ashelhorse
Session Name: human-chimp fusion relationship
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 104
|
1758009600 |
104 |
 |
Description: hg38 chr2 expanded synteny
Author: allygarrett
Session Name: hg38 chr2 expanded synteny
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 36
|
1758009600 |
36 |
 |
Description: Genome Browser Assignment
Author: SamiraL
Session Name: Ridley Ch2 Species Chimps & other 6
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 26
|
1758009600 |
26 |
 |
Description: hg38 vs primates chr2 9/16/25 Madison Tesnow
Author: MadisonTesnow
Session Name: hg38 vs primates chr2 9/16/25 Madison Tesnow
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 14
|
1758009600 |
14 |
 |
Description: 7 primate comparison to human chr2
Author: averywitt
Session Name: chr2 compared to 7 primate AW
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 14
|
1758009600 |
14 |
 |
Description: Comparison of human chromosome 2 with chromosomes from bonobos, chimpanzees, gorillas, orangutans, proboscis monkeys, and golden snub-nosed monkeys.
Author: messicbr
Session Name: Human-Primate Comparison
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 14
|
1758009600 |
14 |
 |
Description: more chromosome comparison
Author: lgunter15
Session Name: figure2_chimpChr2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 11
|
1758009600 |
11 |
 |
Description: human genome, ridley ch2
Author: lgunter15
Session Name: figure1_chimpChr2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 14
|
1758009600 |
14 |
 |
Description: In class Ridley Ch2 Fig. 2.
Author: LucaDooley
Session Name: hg38 Chimp Fig 2.
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 10
|
1758009600 |
10 |
 |
Description: Genome Browser assignment
Author: SamiraL
Session Name: Ridley Ch2 Species
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 13
|
1758009600 |
13 |
 |
Description: human chr2 vs that of other primates
Author: Elizabeth Callis
Session Name: hg38 human-chimp chr2 with other primates
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 17
|
1758009600 |
17 |
 |
Description: Human chromosome 2 compared to 7 primates
Author: Caesaram
Session Name: hg38 chr2 primates
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 11
|
1758009600 |
11 |
 |
Description: Conservation of chromosome 2 between humans and chimps
Author: Caesaram
Session Name: hg38 chr 2 chimp
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 10
|
1758009600 |
10 |
 |
Description: In class Ridley Ch2 assignment.
Author: LucaDooley
Session Name: hg38 Chimp Fig 1.
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 10
|
1758009600 |
10 |
 |
Description: 77
Author: alexandratrias
Session Name: hg38 7 primates.2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 15
|
1758009600 |
15 |
 |
Description: Fall25_Synteny_Group6_BIO581_Fig2
Author: sronalds
Session Name: Fall25_Synteny_Group6_BIO581_Fig2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 10
|
1758009600 |
10 |
 |
Description: 3
Author: alexandratrias
Session Name: hg38 chimp3
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 11
|
1758009600 |
11 |
 |
Description: hg38 chr3 chimp synteny
Author: allygarrett
Session Name: hg38 chr3 chimp synteny
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 16
|
1758009600 |
16 |
 |
Description: hg38 vs chimp chr2 9/16/25 Madison Tesnow
Author: MadisonTesnow
Session Name: hg38 vs chimp chr2 9/16/25 Madison Tesnow
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 9
|
1758009600 |
9 |
 |
Description: Fall25_Synteny_Group6_BIO581_Fig1
Author: sronalds
Session Name: Fall25_Synteny_Group6_BIO581_Fig1
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 9
|
1758009600 |
9 |
 |
Description: chromosome 2 with added species for comparison
Author: katelynnmenjivar
Session Name: chromosome 2 in comparison to other species
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 9
|
1758009600 |
9 |
 |
Description: 2
Author: alexandratrias
Session Name: hg38 chimp2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 10
|
1758009600 |
10 |
 |
Description: chromosome 2 with highlighted region
Author: katelynnmenjivar
Session Name: chromosome 2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 22
|
1758009600 |
22 |
 |
Description: Human Chromosome 3 in comparison to chimp chromosomes
Author: messicbr
Session Name: Human-Chimp Comparison
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 9
|
1758009600 |
9 |
 |
Description: human chr2 vs chimp
Author: Elizabeth Callis
Session Name: hg38 human v chimp chr2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 10
|
1758009600 |
10 |
 |
Description: 7
Author: alexandratrias
Session Name: hg38: 7 primates
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 10
|
1758009600 |
10 |
 |
Description: chr 2 kaya w
Author: Will42ka
Session Name: hg38 7 species human chr2 fusion
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 19
|
1758009600 |
19 |
 |
Description: 7 primate genomes comparisons
Author: Delleeb
Session Name: hg38 chromosome 2 7 genomes
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 14
|
1758009600 |
14 |
 |
Description: chr 2
Author: Will42ka
Session Name: hg38 chimp human chr2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 13
|
1758009600 |
13 |
 |
Description: Human CRX gene mutations
Author: allygarrett
Session Name: hg38 CRX gene
Genome Assembly: hg38
Creation Date: 2025-09-11
Views: 12
|
1757577600 |
12 |
 |
Description: CRX to see annotations and mutatuions
Author: lgunter15
Session Name: CRX
Genome Assembly: hg38
Creation Date: 2025-09-11
Views: 16
|
1757577600 |
16 |
 |
Description: for in class assignment
Author: averywitt
Session Name: Avery CRX practice
Genome Assembly: hg38
Creation Date: 2025-09-11
Views: 16
|
1757577600 |
16 |
 |
Description: CRX gene isoforms
Author: sronalds
Session Name: Fall25_CRX gene isoforms_BIO581_Group6
Genome Assembly: hg38
Creation Date: 2025-09-11
Views: 15
|
1757577600 |
15 |
 |
Description: CRX gene w uniprot
Author: Will42ka
Session Name: hg38 CRX
Genome Assembly: hg38
Creation Date: 2025-09-11
Views: 14
|
1757577600 |
14 |
 |
Description: CRX gene and mutations
Author: Caesaram
Session Name: Hg38 Crx 9/11
Genome Assembly: hg38
Creation Date: 2025-09-11
Views: 11
|
1757577600 |
11 |
 |
Description: bitter tasting alleles
Author: lgunter15
Session Name: TAS2R38
Genome Assembly: hg19
Creation Date: 2025-09-09
Views: 47
|
1757404800 |
47 |
 |
Description: SNP with OMIM in hg19
Author: Elizabeth Callis
Session Name: hg19 SNP 1
Genome Assembly: hg19
Creation Date: 2025-09-09
Views: 24
|
1757404800 |
24 |
 |
Description: hg19 TAS2R38
Author: Delleeb
Session Name: hg19 TAS2R38 ED
Genome Assembly: hg19
Creation Date: 2025-09-09
Views: 10
|
1757404800 |
10 |
 |
Description: hg19 TAS2R38
Author: allygarrett
Session Name: hg19 TAS2R38
Genome Assembly: hg19
Creation Date: 2025-09-09
Views: 12
|
1757404800 |
12 |
 |
Description: TAS2R38
Author: sronalds
Session Name: Fall25_TAS2R38_BIO581_Group6
Genome Assembly: hg19
Creation Date: 2025-09-09
Views: 14
|
1757404800 |
14 |
 |
Description: SNP in hg19
Author: Elizabeth Callis
Session Name: hg19 SNP
Genome Assembly: hg19
Creation Date: 2025-09-09
Views: 11
|
1757404800 |
11 |
 |
Description: Genomics
Author: sronalds
Session Name: Fall25_Dinoflagellates_Group6
Genome Assembly: hg38
Creation Date: 2025-09-15
Views: 14
|
1757923200 |
14 |
 |
Description: Fall25 Yeast BIO481 group2
Author: allygarrett
Session Name: Fall25 Yeast BIO481 group2
Genome Assembly: sacCer3
Creation Date: 2025-09-03
Views: 45
|
1756886400 |
45 |
 |
Description: Rho gene
Author: LucaDooley
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 15
|
1756800000 |
15 |
 |
Description: hghlights and pack view things
Author: Will42ka
Session Name: hg19 rho w highlights
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 34
|
1756800000 |
34 |
 |
Description: hg19 rho
Author: Will42ka
Session Name: hg19 rho
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 9
|
1756800000 |
9 |
 |
Description: Conservation of RHO exons 2 and 3 compared with mouse, chicken, and zebrafish
Author: Caesaram
Session Name: hg19 RHO Cons
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 10
|
1756800000 |
10 |
 |
Description: RHO gene quiz #2
Author: ashelhorse
Session Name: hg19 RHO #2
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 10
|
1756800000 |
10 |
 |
Description: Human RHO gene with OMIM track
Author: Caesaram
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 19
|
1756800000 |
19 |
 |
Description: hg19 RHO quiz 2 Ally Garrett
Author: allygarrett
Session Name: hg19 RHO Quiz 2
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 11
|
1756800000 |
11 |
 |
Description: hg19 RHO
Author: allygarrett
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 12
|
1756800000 |
12 |
 |
Description: Quiz 2
Author: sronalds
Session Name: hg19_Q2
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 12
|
1756800000 |
12 |
 |
Description: hg19 RHO
Author: sronalds
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-09-02
Views: 10
|
1756800000 |
10 |
 |
Description: RHO gene with OMIM Alleles
Author: ashelhorse
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-09-01
Views: 13
|
1756713600 |
13 |
 |
Description: Quiz 2
Author: SamiraL
Session Name: HG19 RHO QUIZ 2
Genome Assembly: hg19
Creation Date: 2025-09-01
Views: 15
|
1756713600 |
15 |
 |
Description: pdf v
Author: zshiff
Session Name: hg19 pdf v
Genome Assembly: hg19
Creation Date: 2025-09-01
Views: 21
|
1756713600 |
21 |
 |
Description: hg19 RHO
Author: zshiff
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-09-01
Views: 18
|
1756713600 |
18 |
 |
Description: RHO gene for quiz 2
Author: averywitt
Session Name: hg19 RHO quiz 2
Genome Assembly: hg19
Creation Date: 2025-09-01
Views: 10
|
1756713600 |
10 |
 |
Description: hg19 RHO gene for quiz 1
Author: averywitt
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-09-01
Views: 17
|
1756713600 |
17 |
 |
Description: Rhodopsin gene view with highlights on exons 2 and 3
Author: messicbr
Session Name: hg19 v2
Genome Assembly: hg19
Creation Date: 2025-09-01
Views: 14
|
1756713600 |
14 |
 |
Description: View of the Rhodopsin gene with OMIM track
Author: messicbr
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-09-01
Views: 14
|
1756713600 |
14 |
 |
Description: quiz 2 submission
Author: lgunter15
Session Name: hg19_quiz2
Genome Assembly: hg19
Creation Date: 2025-08-31
Views: 13
|
1756627200 |
13 |
 |
Description: hg19 RHO part 2 for quiz #2
Author: katelynnmenjivar
Session Name: hg19 RHO Part 2 KM
Genome Assembly: hg19
Creation Date: 2025-08-31
Views: 11
|
1756627200 |
11 |
 |
Description: RHO hg19 done by Lilymae Gunter for Quiz assignment
Author: lgunter15
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-08-31
Views: 12
|
1756627200 |
12 |
 |
Description: h19 RHO quiz 1 practice
Author: katelynnmenjivar
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-08-31
Views: 14
|
1756627200 |
14 |
 |
Description: quiz 1
Author: alexandratrias
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2025-08-31
Views: 13
|
1756627200 |
13 |
 |
Description: Fall25 Yeast BIO481 group#2
Author: MadisonTesnow
Session Name: Fall25 Yeast BIO481 group#2
Genome Assembly: sacCer3
Creation Date: 2025-08-30
Views: 14
|
1756540800 |
14 |
 |
Description: The yeast activity performed in class
Author: annesophierosa
Session Name: CGEMS yeast AR test
Genome Assembly: sacCer3
Creation Date: 2025-08-28
Views: 12
|
1756368000 |
12 |
 |
Description: Genomics class
Author: sronalds
Session Name: Fall25 MouseBIO581group6
Genome Assembly: mm10
Creation Date: 2025-08-28
Views: 10
|
1756368000 |
10 |
 |
Description: Yeast Genomic Browser Activity
Author: SamiraL
Session Name: CGEMS Worm SL Test
Genome Assembly: sacCer3
Creation Date: 2025-08-28
Views: 16
|
1756368000 |
16 |
 |
Description: bio 481-worm
Author: katelynnmenjivar
Session Name: CGEMS Worm KM test
Genome Assembly: ce11
Creation Date: 2025-08-28
Views: 16
|
1756368000 |
16 |
 |
Description: C. elegans EDG2
Author: ashelhorse
Session Name: Fall25 Yeast BIO481 group#2
Genome Assembly: sacCer3
Creation Date: 2025-08-28
Views: 13
|
1756368000 |
13 |
 |
Description: S. cerevisiae yeast genome browser day 1
Author: SamiraL
Session Name: Fall25 Yeast BIO481 group 1
Genome Assembly: sacCer3
Creation Date: 2025-08-28
Views: 10
|
1756368000 |
10 |
 |
Description: class assignment Group 1 Yeast/Lilymae Gunter
Author: lgunter15
Session Name: Fall25 Yeast BIO481 group1
Genome Assembly: sacCer3
Creation Date: 2025-08-28
Views: 11
|
1756368000 |
11 |
 |
Description: class work group 1 yeast AW
Author: averywitt
Session Name: Fall25 Yeast BIO481 group#1
Genome Assembly: sacCer3
Creation Date: 2025-08-28
Views: 8
|
1756368000 |
8 |
 |
Description: Fall25 MouseBIO481 group5
Author: zshiff
Session Name: CGEMS Mouse ZKS Test
Genome Assembly: mm39
Creation Date: 2025-08-28
Views: 14
|
1756368000 |
14 |
 |
Description: BIO481 C. elegans genome assignment
Author: messicbr
Session Name: Fall2025 Worm BIO481 Group #4
Genome Assembly: ce11
Creation Date: 2025-08-28
Views: 11
|
1756368000 |
11 |
 |
Description: Fall25 Worm BIO481 group#3
Author: Elizabeth Callis
Session Name: Fall25 Worm BIO481 group#3
Genome Assembly: ce11
Creation Date: 2025-08-28
Views: 15
|
1756368000 |
15 |
 |
Description: Fall25 C. elegans Bio481 group3
Author: oliviakloc
Session Name: Fall25 Worm Bio481 group3
Genome Assembly: ce11
Creation Date: 2025-08-28
Views: 19
|
1756368000 |
19 |
 |
Description: Fall 2025 C. elegans Bio 481 Group 3
Author: Elizabeth McLaughlin
Session Name: Fall 2025 Worm Bio 481 Group 3
Genome Assembly: ce11
Creation Date: 2025-08-28
Views: 14
|
1756368000 |
14 |
 |
Description: Worm group #3
Author: Hailandon
Session Name: Fall25 Worm Bio481 #3
Genome Assembly: ce11
Creation Date: 2025-08-28
Views: 11
|
1756368000 |
11 |
 |
Description: Kaya C. elegans
Author: Will42ka
Session Name: FALL25 KW Worm Bio481 group4
Genome Assembly: ce11
Creation Date: 2025-08-28
Views: 13
|
1756368000 |
13 |
 |
Description: MOuse mm39
Author: Caesaram
Session Name: Fall25 MouseBIO481 group #6
Genome Assembly: mm39
Creation Date: 2025-08-28
Views: 15
|
1756368000 |
15 |
 |
Description: C. elegans ICD-1
Author: Delleeb
Session Name: Emily Delle - Fall25 C.elegans BIO481/581 group4
Genome Assembly: ce11
Creation Date: 2025-08-28
Views: 23
|
1756368000 |
23 |
 |
Description: Session on Hg38 to interpret SV variants
Author: csauve
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2024-07-23
Views: 199
|
1721721600 |
199 |
 |
Description: OG is the predominant product of oxidative damage in the genome and has been shown to play an important regulatory role under pathological conditions such as cancer and aging. GOAL-MAP enabled profiling of OG landscape of mouse cells.
Author: KAI_LI
Session Name: Mus_cell_line_OG_map
Genome Assembly: mm10
Creation Date: 2025-10-28
Views: 15
|
1761638400 |
15 |
 |
Description: OG is the predominant product of oxidative damage in the genome and has been shown to play an important regulatory role under pathological conditions such as cancer and aging. GOAL-MAP enabled profiling of OG landscape of mouse and human cells, revealing the differential distribution of OG modifications and their potential regulatory functions in gene expression.
Author: KAI_LI
Session Name: Huma cell line OG map
Genome Assembly: hg38
Creation Date: 2025-10-28
Views: 25
|
1761638400 |
25 |
 |
Description: CORSIV Regions discovered in White and Black WGBS data
Author: gunaseka
Session Name: Black_White_CoRSIVs
Genome Assembly: hg38
Creation Date: 2025-08-20
Views: 51
|
1755676800 |
51 |
 |
Description: MCB197 Harvard
Author: amandajoyward
Session Name: MCB197-2A-WGBS
Genome Assembly: hg38
Creation Date: 2025-08-18
Views: 62
|
1755504000 |
62 |
 |
Description: HEK293T_ChIP_sgRNA
Author: lkhwan0929
Session Name: HEK293T_ChIP_sgRNA
Genome Assembly: hg38
Creation Date: 2025-08-21
Views: 49
|
1755763200 |
49 |
 |
Description: A UCSC session with public ATAC seq data
Author: JaviML98
Session Name: ATAC-seqData
Genome Assembly: hg19
Creation Date: 2025-08-11
Views: 17
|
1754899200 |
17 |
 |
Description: Zoomed in Exon 1 CAG x7 repeats
Author: Caesaram
Session Name: hg38 HTT CAG
Genome Assembly: hg38
Creation Date: 2025-10-07
Views: 10
|
1759824000 |
10 |
 |
Description: HP1a_DamID in central brain from Drosophila 3rd instar larvae upon Ago2 mutation or Lam-KD in neurons.
Author: Shevelyov
Session Name: HP1a_DamID in brain upon Ago2mut or Lam-KD in neurons
Genome Assembly: dm3
Creation Date: 2025-08-05
Views: 14
|
1754380800 |
14 |
 |
Description: Trout Peak chipSeaq, ATACSeaq
Functional annotation of regulatory elements in rainbow trout uncovers roles of the epigenome in genetic selection and genome evolution
Author: rafet
Session Name: GCF_013265735.2G
Genome Assembly: hub_6326273_GCF_013265735.2
Creation Date: 2025-09-15
Views: 19
|
1757923200 |
19 |
 |
Description: H4panAc changes in S2 cells upon TSA or curcumin treatments, and upon GFP-HAT-Lamin expression
Author: Shevelyov
Session Name: H4panAc changes in S2 upon TSA and curcumin treatments or GFP-HAT-Lamin expression
Genome Assembly: dm3
Creation Date: 2025-07-28
Views: 20
|
1753689600 |
20 |
 |
Description: hg38 chromosome 2 with chimp genome
Author: Delleeb
Session Name: hg38 chromosome 2
Genome Assembly: hg38
Creation Date: 2025-09-16
Views: 27
|
1758009600 |
27 |
 |
Description: FigS6 modified from this session.
Author: 429035671
Session Name: Pei_et_al_2025_figS6_UCSC_track
Genome Assembly: hg19
Creation Date: 2025-07-11
Views: 141
|
1752220800 |
141 |
 |
Description: UCSC track for FigS1
Author: 429035671
Session Name: Pei_et_al_2025_figS1_UCSC_track
Genome Assembly: hg19
Creation Date: 2025-07-11
Views: 136
|
1752220800 |
136 |
 |
Description: OG is the predominant product of oxidative damage in the genome and has been shown to play an important regulatory role under pathological conditions such as cancer and aging. Our studies developed a new genome-wide single-base OG sequencing technology and provided a key resource for clarifying the role of OG in aging.
Author: KAI_LI
Session Name: Mouse_tissue_OG_landscape
Genome Assembly: mm10
Creation Date: 2025-10-28
Views: 21
|
1761638400 |
21 |
 |
Description: T2D_elizabeth
Author: Elizabeth Alatorre
Session Name: T2D_elizabeth
Genome Assembly: hg38
Creation Date: 2025-07-07
Views: 209
|
1751875200 |
209 |
 |
Description: AD Dark Microproteome
Author: brmiller
Session Name: AD Dark Microproteome
Genome Assembly: hg38
Creation Date: 2025-04-03
Views: 318
|
1743667200 |
318 |
 |
Description: Prenatal fam study: de novo 1p12p11.2
Author: casey.brewer
Session Name: 1p12p11.2 dn
Genome Assembly: hg19
Creation Date: 2025-06-30
Views: 271
|
1751270400 |
271 |
 |
Description: Multiregion view of 66 MANE Plus Clinical transcripts
Author: AlistairP
Session Name: MANE_PLUS_CLINICAL
Genome Assembly: hg38
Creation Date: 2025-06-25
Views: 138
|
1750838400 |
138 |
 |
Description: This is the session showing the location and expression of the new junction that might cause frame shift.
Author: dandan_0909
Session Name: hg19_version
Genome Assembly: hg19
Creation Date: 2025-05-28
Views: 738
|
1748419200 |
738 |
 |
Description: This is the session showing the location and expression of the new junction and NR junctions
Author: dandan_0909
Session Name: hg19_all_new_nr
Genome Assembly: hg19
Creation Date: 2025-06-09
Views: 345
|
1749456000 |
345 |
 |
Description: This is the session showing the location and expression of the new junction and NR junctions
Author: dandan_0909
Session Name: hg38_all_new_nr
Genome Assembly: hg38
Creation Date: 2025-06-09
Views: 217
|
1749456000 |
217 |
 |
Description: data from doi: 10.1101/gr.218602.116. Epub
Author: sansamcl
Session Name: danRer10_RT_chr4
Genome Assembly: danRer10
Creation Date: 2025-05-29
Views: 208
|
1748505600 |
208 |
 |
Description: HEK293T cell lines engineered to express Empty Vector Control (EV_783) or mutant U2 snRNA (MUT_C28U or MUT_UCA). For each cell line there are 3 tracks for the 3 biological replicates, and are ordered and colored based on the cell line. Bigwig files were created for the forward and reverse reads using deeptools (v3.5.6) and read counts were normalized using the inverse of the DEseq2 size factors.
Author: mstevers
Session Name: U2muttracks_groupedcolor
Genome Assembly: hg38
Creation Date: 2025-05-14
Views: 54
|
1747209600 |
54 |
 |
Description: Public session with custom track
Author: MariaDeluca
Session Name: hg38_wtest.bed
Genome Assembly: hg38
Creation Date: 2025-11-30
Views: 14
|
1764489600 |
14 |
 |
Description: things
Author: apehkone
Session Name: MOMIC2_LNCAP
Genome Assembly: hg19
Creation Date: 2025-05-11
Views: 497
|
1746950400 |
497 |
 |
Description: The UCSC session data for the Long-read Genome Sequencing.
Author: Yuki_INOUE
Session Name: Genome-seq Session
Genome Assembly: mm39
Creation Date: 2025-05-05
Views: 449
|
1746432000 |
449 |
 |
Description: My whole genome sequencing data from October 2024
Author: becca88
Session Name: Rebecca WGS
Genome Assembly: hg38
Creation Date: 2025-04-16
Views: 992
|
1744790400 |
992 |
 |
Description: Runx3 Cut&Run on MNK3 Cells
Author: pattonjt
Session Name: Run3_CR_MNK3
Genome Assembly: mm10
Creation Date: 2025-04-15
Views: 436
|
1744704000 |
436 |
 |
Description: retina
Author: ALLDEREP
Session Name: mm9MouseRETINA2025
Genome Assembly: mm9
Creation Date: 2025-04-08
Views: 525
|
1744099200 |
525 |
 |
Description: hg38
Used for primer design to show the variants from dbSNP155, Gnomad, SNPedia and 1000 genome
Author: vvenugopal
Session Name: hg38_primer_design
Genome Assembly: hg38
Creation Date: 2025-04-08
Views: 361
|
1744099200 |
361 |
 |
Description: e
Author: ALLDEREP
Session Name: hg38MAPTGENE
Genome Assembly: hg38
Creation Date: 2025-04-03
Views: 209
|
1743667200 |
209 |
 |
Description: e
Author: ALLDEREP
Session Name: hg19RHOCUSTOM
Genome Assembly: hg19
Creation Date: 2025-04-03
Views: 182
|
1743667200 |
182 |
 |
Description: The CLOCK gene that regulates circadian rhythms, using human genome GRCh38/hg38.
Author: sjacques
Session Name: circadian_for_submission
Genome Assembly: hg38
Creation Date: 2025-04-03
Views: 184
|
1743667200 |
184 |
 |
Description: dna
Author: Jessra9
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2025-04-11
Views: 195
|
1744358400 |
195 |
 |
Description: RNA-seq, ChIP-seq and ATAC-seq data in the study "The SAS Chromatin-Remodeling Complex Mediates Inflorescence-Specific Chromatin Accessibility for Transcription Factor Binding".
Author: GuoJ
Session Name: NAR_SAS_sdl_ifl_all_data
Genome Assembly: hub_2660163_GCF_000001735.4
Creation Date: 2025-03-29
Views: 214
|
1743235200 |
214 |
 |
Description: diseño primers
Author: Paulabm
Session Name: diseño primers
Genome Assembly: hg19
Creation Date: 2025-03-25
Views: 50
|
1742889600 |
50 |
 |
Description: Exonic enhancers (EEs) are a newly characterized class of dual-function regulatory elements that reside in protein-coding regions yet retain bona fide enhancer activity. In our study, we integrated transcription factor (TF) binding profiles (ReMap), chromatin accessibility data (DNase, ATAC), and high-throughput reporter assays (STARR-seq, luciferase) to show that many exons can act as cis-regulatory modules. These EEs display hallmark epigenomic signatures, often form long-range interactions with promoters, and are sensitive to both nonsynonymous and synonymous mutations. Functional assays, including CRISPR-based silencing, demonstrate their involvement in regulating host and distal genes. Large-scale cancer genomics analyses reveal that EE variants are associated with dysregulated gene expression and clinical outcomes, underscoring their importance in disease. Evolutionary comparisons further indicate that EEs combine deep sequence conservation with lineage-specific innovation, revealing how coding regions can simultaneously encode protein domains and regulatory activities.
This UCSC public session provides direct visualization of these findings, featuring our custom track hub: https://remap.univ-amu.fr/storage/public/hubEE/hub.txt.
Researchers can explore TF occupancy, open chromatin marks, STARR-seq signals, and EE annotations genome-wide, offering a comprehensive resource to investigate how exons function as both protein-coding and cis-regulatory elements.
Author: Benoit Ballester
Session Name: hg38_ExonEnhancers_gnomAD
Genome Assembly: hg38
Creation Date: 2025-03-21
Views: 526
|
1742544000 |
526 |
 |
Description: Exonic enhancers (EEs) are a newly characterized class of dual-function regulatory elements that reside in protein-coding regions yet retain bona fide enhancer activity. In our study, we integrated transcription factor (TF) binding profiles (ReMap), chromatin accessibility data (DNase, ATAC), and high-throughput reporter assays (STARR-seq, luciferase) to show that many exons can act as cis-regulatory modules. These EEs display hallmark epigenomic signatures, often form long-range interactions with promoters, and are sensitive to both nonsynonymous and synonymous mutations. Functional assays, including CRISPR-based silencing, demonstrate their involvement in regulating host and distal genes. Large-scale cancer genomics analyses reveal that EE variants are associated with dysregulated gene expression and clinical outcomes, underscoring their importance in disease. Evolutionary comparisons further indicate that EEs combine deep sequence conservation with lineage-specific innovation, revealing how coding regions can simultaneously encode protein domains and regulatory activities.
This UCSC public session provides direct visualization of these findings, featuring our custom track hub: https://remap.univ-amu.fr/storage/public/hubEE/hub.txt.
Researchers can explore TF occupancy, open chromatin marks, STARR-seq signals, and EE annotations genome-wide, offering a comprehensive resource to investigate how exons function as both protein-coding and cis-regulatory elements.
Author: Benoit Ballester
Session Name: hg19_ExonEnhancers_TCGA
Genome Assembly: hg19
Creation Date: 2025-03-21
Views: 428
|
1742544000 |
428 |
 |
Description: This public session offers access to a track hub containing 25,000 predicted forebrain enhancers within the hg19 human genome assembly. These enhancers were identified through a DNA-sequence-based prediction pipeline that integrates tissue-specific transcription factor occupancy patterns. Essential for regulating gene expression during brain development, these enhancers were further validated using chromatin marks, DNase hypersensitivity data, GWAS-based SNP data, and in vivo zebrafish models (https://doi.org/10.1002/1873-3468.15030).
Additionally, this session includes an extra track for a subset of 2,614 predicted forebrain enhancers, representing a portion of the full set. This subset specifically reveals the binding patterns of the HES5-FOXP2-GATA3 triad, with FOXP2 binding positioned between HES5 and GATA3, helping define the forebrain transcription factor binding syntax (https://doi.org/10.1242/bio.061751).
Together, the forebrain enhancer data presented in this public track hub provides a valuable resource for investigating the genetic regulatory mechanisms underlying brain development and related diseases. It also offers a platform for exploring the role of these predicted enhancers in mammalian and primate brain evolution.
Author: abbasiam
Session Name: hg19_Predicted human forebrain enhancers_hg19
Genome Assembly: hg19
Creation Date: 2025-03-20
Views: 387
|
1742457600 |
387 |
 |
Description: This session give an overview of the human CHD1 gene encoding the clinically relevant E-Cadherin/Cadherin-1 protein. Disease susceptibility is associated with variants in CDH-1 including gastric cancer and lobular breast cancer. Data tracks demonstrating disease-associated alleles, protein domains, and conservation have been included.
Author: renkejhsph
Session Name: hg38 CDH1 gene
Genome Assembly: hg38
Creation Date: 2025-03-20
Views: 180
|
1742457600 |
180 |
 |
Description: The Kidney Disease Genetic Scorecard is a multitude of multi-omics datasets for the annotation of regulatory variants, encompassing 32 kinds of genetic evidence including ASE, bASA, snASA, Open4Gene links, and coding variants. This strategy enabled us to prioritize 601 genes targeted by both regulatory variants and coding variants, including 161 genes with convergence of two kinds of variants. The details information is available in Liu et al., Science 2025 (https://www.science.org/doi/full/10.1126/science.adp4753).
Author: hongbo919
Session Name: Kidney.Disease.Genetic.Scorecard
Genome Assembly: hg19
Creation Date: 2024-07-03
Views: 2769
|
1719993600 |
2769 |
 |
Description: 1
ZFP36L2 (zinc finger protein 36 like 2, C3H type-ZFP) is an RNA-binding protein targeting
transcripts rich in adenine-uridine elements (AREs). Previous transcriptomic analysis
suggested that ZFP36L2 displays a distinct transcript preference, depending on the tissue
of expression. However, this analysis was restricted to a few tissues. Here, we collected
RNA-seq data in six tissues and detected a remarkable transcript selectivity. Given that
ZFP36L2 accelerates the degradation of specific ARE-transcripts upon binding, we
obtained differential expression transcriptomic data on a Zfp36l2 knock-out mouse model
to delve into the mechanisms governing this tissue-specific targeting. Transcriptomic
analyzes of up regulated ARE-transcripts in lung, liver, bone marrow, spleen, kidney, and
ovary of the Zfp36l2-deficient mouse confirmed that there is high tissue preference in
ZFP36L2 targets. We observed only one common up regulated gene, Apol11b, among
these six different tissues. However, we do observe common trends, specifically an
enrichment in protein coding genes in the up regulated genes, consistent with these RBP
primarily targeting genes on their 3’ UTRs. Interestingly, we observed a significant
increase in the proportion of IG (immunoglobulin) genes being up regulated. We further
performed eCLIP (Enhanced Cross-Linking&ImmunoPreciptation) on a mouse cell line,
MLTC-1 cells, to identify direct binding sites of ZFP36L2. AU-Rich Element score
(AREscore) analysis revealed enrichment in both up regulated genes and eCLIP peaks,
although some differences were observed in flanking residue composition. Our findings
provide new insights into the intricate regulatory network orchestrated by ZFP36L2,
opening avenues for exploring its potential roles in different tissues.
Author: gsgeorge
Session Name: Zfp36l2_mm10_eclip_peaks
Genome Assembly: mm10
Creation Date: 2025-03-27
Views: 192
|
1743062400 |
192 |
 |
Description: Module 08 test.bed
Author: froemming.m
Session Name: froemming.m_mod_8
Genome Assembly: hg38
Creation Date: 2025-03-10
Views: 654
|
1741593600 |
654 |
 |
Description: For CNV analysis
Author: HGSalk
Session Name: CNV
Genome Assembly: hg19
Creation Date: 2025-03-06
Views: 373
|
1741248000 |
373 |
 |
Description: mm39 Blue opsin F2-R2 amplicon (#386-387) for BS seq
Author: renkejhsph
Session Name: mm39 Blue opsin F2-R2 amplicon (#386-387)
Genome Assembly: mm39
Creation Date: 2025-03-04
Views: 447
|
1741075200 |
447 |
 |
Description: It is tracked by samples and treatment groups. Some differentially accessible peaks highlighted.Custom gtf file with transcript ID, gene names, gene ID
Author: Pengxin Yang
Session Name: pig_BCG_ATAC_filtered_by_sample_and_merged_custom_gtf
Genome Assembly: susScr11
Creation Date: 2025-03-03
Views: 352
|
1740988800 |
352 |
 |
Description: NSD1 truncating variants
Author: michel.guipponi@hug.ch
Session Name: hg38_NSD1_Truncating
Genome Assembly: hg38
Creation Date: 2025-03-02
Views: 305
|
1740902400 |
305 |
 |
Description: Why don't humans have tails?
Author: michel.guipponi@hug.ch
Session Name: hg38_TBXT
Genome Assembly: hg38
Creation Date: 2025-03-02
Views: 178
|
1740902400 |
178 |
 |
Description: NSD1 missense hot-spot
Author: michel.guipponi@hug.ch
Session Name: hg38_NSD1_missense
Genome Assembly: hg38
Creation Date: 2025-03-02
Views: 211
|
1740902400 |
211 |
 |
Description: E2F1 binding site on genome depending on SETD6
Author: Gizem
Session Name: E2F1 chip seq
Genome Assembly: hg38
Creation Date: 2025-02-27
Views: 263
|
1740643200 |
263 |
 |
Description: It has tracks by samples and by treatment groups
Author: Pengxin Yang
Session Name: pig_BCG_ATAC_filtered_by_sample_and_merged
Genome Assembly: susScr11
Creation Date: 2025-02-27
Views: 171
|
1740643200 |
171 |
 |
Description: ALDH2 and alcohol intolerance in East Asian populations
Author: michel.guipponi@hug.ch
Session Name: hg38_ALDH2
Genome Assembly: hg38
Creation Date: 2025-03-02
Views: 223
|
1740902400 |
223 |
 |
Description: Each bam file was filtered in the identified 92226 peaks.
Author: Pengxin Yang
Session Name: pig_BCG_ATAC_filtered_by_sample
Genome Assembly: susScr11
Creation Date: 2025-02-26
Views: 277
|
1740556800 |
277 |
 |
Description: mTEClo vs mTEChi anti-P53 CUT&RUN from Gamble et al. 2025
Author: ngamble
Session Name: antiP53_CUTRUN_mTECs
Genome Assembly: mm10
Creation Date: 2025-02-25
Views: 287
|
1740470400 |
287 |
 |
Description: A few DAPs highlighted
Author: Pengxin Yang
Session Name: pig_BCG_project_ATAC _highlight
Genome Assembly: susScr11
Creation Date: 2025-02-17
Views: 539
|
1739779200 |
539 |
 |
Description: peaks visualization for pig BCG project. A few DAPs were highlighted
Author: Pengxin Yang
Session Name: pig_BCG_project_ATAC
Genome Assembly: susScr11
Creation Date: 2025-02-17
Views: 354
|
1739779200 |
354 |
 |
Description: Machine Learning Prediction of Non-Coding Variant Impact in Cell-Class-Specific Human Retinal Cis-Regulatory Elements
Leah S. VandenBosch 1 and Timothy J. Cherry 1,2,3†
1 Center for Developmental Biology and Regenerative Medicine, Seattle Children's Research Institute, Seattle, WA, USA.
2 University of Washington Department of Pediatrics, Seattle, Washington, USA.
3 Brotman Baty Institute for Precision Medicine, Seattle, Washington, USA.
† Corresponding Author: timothy.cherry@seattlechildrens.org
This public session provides access to a track hub of Variant Impact Prediction (VIP) scores for 39,437 candidate human retinal cis-regulatory elements (CREs) in 18 different cell types and developmental stages from the adult and developing human retina.
Non-coding variants in cis-regulatory elements (CREs) like promoters and enhancers contribute to inherited retinal diseases (IRDs) however regulatory variants are typically challenging to interpret and prioritize for follow up studies. To improve identification of variants of interest, we implemented machine learning using a gapped k-mer support vector machine approach (GKM-SVM/deltaSVM) trained on single nucleus ATAC-seq data from specific cell classes and developmental states in the developing and mature human retina. These models demonstrate accuracy over 90% and a high degree of cell class specificity. Variant Impact Prediction Scores (VIPS) nominate specific sequences within candidate CREs, including putative transcription factor (TF) binding motifs, that would likely alter CRE function if mutated. This analysis demonstrates the capacity for singe nucleus-epigenomic data to predict the impact of non-coding sequence variants. It is our hope that these predicted impact scores can assist in identifying and interpreting variants of interest within human retinal cis-regulatory elements.
Author: CherryLab
Session Name: csVISIONS_TrackHub
Genome Assembly: hg38
Creation Date: 2025-02-16
Views: 654
|
1739692800 |
654 |
 |
Description: rho
Author: ALLDEREP
Session Name: hg19rHONEW
Genome Assembly: hg19
Creation Date: 2025-01-28
Views: 526
|
1738051200 |
526 |
 |
Description: Abigail Hodges
Author: aehodges
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-08-31
Views: 514
|
1725091200 |
514 |
 |
Description: single cell bone marrow analysis
Author: marcel
Session Name: BM_scRNA_PB_ID _paper
Genome Assembly: hg38
Creation Date: 2025-01-10
Views: 188
|
1736496000 |
188 |
 |
Description: mm10_CTCF-CHD7 bigwig signal tracks
Author: jyq_ucsc_2
Session Name: mm10_CTCF-CHD7
Genome Assembly: mm10
Creation Date: 2025-01-08
Views: 720
|
1736323200 |
720 |
 |
Description: Sept 9th UCSC SNP Quiz Session
Author: jdkomine
Session Name: Sept9 UCSC SNP Quiz Session
Genome Assembly: hg19
Creation Date: 2025-09-09
Views: 15
|
1757404800 |
15 |
 |
Description: yes
Author: aryanzandi123
Session Name: Core Chromatin State Window
Genome Assembly: hg38
Creation Date: 2024-12-11
Views: 995
|
1733904000 |
995 |
 |
Description: CTCF and SMC1 ChIP-seq data for Crewe et al submission to NAR
Author: Morgan.Crewe
Session Name: NAR_CTCF_SMC1_Data
Genome Assembly: mm10
Creation Date: 2024-12-10
Views: 928
|
1733817600 |
928 |
 |
Description: Training for visualising transcriptomic data
Author: afrankish
Session Name: longtrec2
Genome Assembly: hg38
Creation Date: 2025-03-02
Views: 246
|
1740902400 |
246 |
 |
Description: 2/2/25
Author: aehodges
Session Name: hg38//CRX gene
Genome Assembly: hg38
Creation Date: 2025-03-02
Views: 160
|
1740902400 |
160 |
 |
Description: A new study in Nature reveals how the absence of a small gene segment due to alternative splicing in CPEB4 leads to the deregulation of 200 genes associated with autism spectrum disorders: https://www.nature.com/articles/s41586-024-08289-w An earlier work by the same authors identified this mRNA as a factor in the autism-like phenotype: https://www.nature.com/articles/s41586-018-0423-5 The work took place at IRB Barcelona: https://www.irbbarcelona.org/en/news/scientific/key-breakthrough-autism-pivotal-role-cpeb4-condensates-revealed: “Our results suggest that even small decreases in the percentage of microexon inclusion can have significant effects. This would explain why some individuals without a gene mutation develop idiopathic autism.” UCSC Genome Browser Session
Author: brianlee
Session Name: CPEB4_GCAAGGACATATGGGCGAAGGAGA
Genome Assembly: hg38
Creation Date: 2024-12-05
Views: 313
|
1733385600 |
313 |
 |
Description: Rearrangements of the 17q24.3 region associated with abnormal phenotypes.
Author: 429035671
Session Name: Pei_et_al_2025_fig5
Genome Assembly: hg19
Creation Date: 2024-12-04
Views: 492
|
1733299200 |
492 |
 |
Description: 12
Author: aryanzandi123
Session Name: Core Chromatin State Window 2.0
Genome Assembly: hg38
Creation Date: 2024-12-11
Views: 265
|
1733904000 |
265 |
 |
Description: te
Author: aryanzandi123
Session Name: Alternate Repressors and Activators Window
Genome Assembly: hg38
Creation Date: 2024-12-11
Views: 297
|
1733904000 |
297 |
 |
Description: 15 stae ChromHMM model of MSC,PC,HC based on murine data and on a human ChromHMM model. (Neu et al. 2024)
Author: manuela.wuelling@uni-due.de
Session Name: murine and humanized ChromHMM model
Genome Assembly: mm39
Creation Date: 2024-11-15
Views: 844
|
1731657600 |
844 |
 |
Description: GFP+ ChIP-Seq data
Author: nimt0001
Session Name: GFP+ ChIP-Seq
Genome Assembly: danRer10
Creation Date: 2024-11-13
Views: 682
|
1731484800 |
682 |
 |
Description: ATAC-Seq from 8 sorted immune cell populations representing the early lymphoid hematopoietic differentiation cascade along the B cell lineage. Cell populations were isolated from healthy adult human bone marrow precursors and peripheral blood (n=13). A total of 78 samples are analyzed. Additional single-cell ATAC-Seq data from 5 healthy adult human bone marrow, recovering HSC, MPP, LMPP, CLP, cycling pro-B, pro-B, cycling pre-B, and pre-B cells, is analyzed.
Author: TransBioLab
Session Name: hg38_Brex
Genome Assembly: hg38
Creation Date: 2024-11-13
Views: 955
|
1731484800 |
955 |
 |
Description: MultiRegion
chrX 1 156040895
chrY 1 57227415
https://www.jax.org/jax-mice-and-services/ipsc
The KOLF2.1J cell line was derived from the HPSI0114i-kolf_2 line through extensive molecular and cellular characterization efforts, including the correction of a pathogenic mutation in ARID2.
https://www.hipsci.org/#/lines/HPSI0114i-kolf_2
Biosample: SAMEA2547615
https://www.ebi.ac.uk/biosamples/samples/SAMEA2547615
subject id SAMEA2398402
https://www.ebi.ac.uk/biosamples/samples/SAMEA2398402
Originating cell line seems to have no chrY data.
CRAM file
/17998_8%235.cram
track name=17998_8%235.cram bigDataUrl=http://ftp.sra.ebi.ac.uk/vol1/run/ERR120/ERR1203439/17998_8%235.cram type=bam
Session: https://genome.ucsc.edu/s/brianlee/cram_KOLF2
Author: brianlee
Session Name: cram_KOLF2
Genome Assembly: hg38
Creation Date: 2024-11-12
Views: 407
|
1731398400 |
407 |
 |
Description: Module 08 Assignment
Author: LouisLiu
Session Name: test
Genome Assembly: hg38
Creation Date: 2024-11-12
Views: 301
|
1731398400 |
301 |
 |
Description: RHO/H1FOO
Author: sofiaterenziani
Session Name: RHO/H1FOO
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 284
|
1730966400 |
284 |
 |
Description: NRL/CPNE6
Author: sofiaterenziani
Session Name: NRL/CPNE6
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 273
|
1730966400 |
273 |
 |
Description: GUCA1B/GUCA1A
Author: sofiaterenziani
Session Name: GUCA1B/GUCA1A
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 277
|
1730966400 |
277 |
 |
Description: PDE6C/RBP4
Author: sofiaterenziani
Session Name: PDE6C/RBP4
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 280
|
1730966400 |
280 |
 |
Description: mm9_NRL_CPNE6
Author: l.g.altizer
Session Name: mm9_NRL_CPNE6
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 292
|
1730966400 |
292 |
 |
Description: mm9_CPNE6_NRL
Author: l.g.altizer
Session Name: mm9_CPNE6_NRL
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 287
|
1730966400 |
287 |
 |
Description: 11/7
Author: aehodges
Session Name: mm9//Nrl gene
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 240
|
1730966400 |
240 |
 |
Description: mm9_RHO_H1FOO
Author: l.g.altizer
Session Name: mm9_RHO_H1FOO
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 267
|
1730966400 |
267 |
 |
Description: 11/7
Author: aehodges
Session Name: mm9//Guac1B gene
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 286
|
1730966400 |
286 |
 |
Description: GUCA1B/GUCA1A
Author: madelinebarlas
Session Name: mm9 GUCA1B/GUCA1A
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 260
|
1730966400 |
260 |
 |
Description: multiomics
Author: diazacex
Session Name: Multiomics_mm9_wt_Nrl-
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 282
|
1730966400 |
282 |
 |
Description: NRL/CPNE6
Author: madelinebarlas
Session Name: mm9 NRL/CPNE6
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 268
|
1730966400 |
268 |
 |
Description: PDE6C/RBP4
Author: madelinebarlas
Session Name: mm9 PDE6C/RBP4
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 260
|
1730966400 |
260 |
 |
Description: Mouse Retina stuff
Author: ALLDEREP
Session Name: mm9MouseRetina
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 302
|
1730966400 |
302 |
 |
Description: PDE6C/RBP4
Author: madisonflorenz
Session Name: PDE6C/RBP4
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 269
|
1730966400 |
269 |
 |
Description: NRL/CPNE6
Author: rchelsea
Session Name: mm9 NRL
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 270
|
1730966400 |
270 |
 |
Description: RHO/H1FOO
Author: rchelsea
Session Name: mm9 RHO
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 278
|
1730966400 |
278 |
 |
Description: GUCA1B/GUCA1A
Author: madisonflorenz
Session Name: GUCA1B/GUCA1A
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 265
|
1730966400 |
265 |
 |
Description: GUCA1B/GUCA1A
Author: rchelsea
Session Name: mm9 GUCA1B
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 261
|
1730966400 |
261 |
 |
Description: 11/7
Author: aehodges
Session Name: mm9//PDE6C gene
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 251
|
1730966400 |
251 |
 |
Description: 11/7
Author: aehodges
Session Name: mm9//RHO
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 269
|
1730966400 |
269 |
 |
Description: PDE6C/RBP4
Author: rchelsea
Session Name: mm9 PDE6C
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 297
|
1730966400 |
297 |
 |
Description: NRL/CPNE6
Author: madisonflorenz
Session Name: NRL/CPNE6
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 280
|
1730966400 |
280 |
 |
Description: mm9
wt CBR
Nrl-
Author: diazacex
Session Name: mm9_BAM_wt CBR_Nrl-
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 255
|
1730966400 |
255 |
 |
Description: RHO/H1FOO
Author: madisonflorenz
Session Name: RHO/H1FOO
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 307
|
1730966400 |
307 |
 |
Description: mm9_GUCA1B_GUCA1A
Author: l.g.altizer
Session Name: mm9_GUCA1B_GUCA1A
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 321
|
1730966400 |
321 |
 |
Description: GB Multiomics custom data tracks - 3
Author: madisonflorenz
Session Name: GB Multiomics custom data tracks - 3
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 277
|
1730966400 |
277 |
 |
Description: mm9_CBR_RNA_seq
Author: l.g.altizer
Session Name: mm9_CBR_RNA_seq
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 280
|
1730966400 |
280 |
 |
Description: wt CBRs
Nrl- CBRs
Author: diazacex
Session Name: mm9_wt CBR_Nrl- CBR
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 289
|
1730966400 |
289 |
 |
Description: CBR regions with extra data
Author: ClaytonFannin
Session Name: mm9 CBR regions with extra data
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 288
|
1730966400 |
288 |
 |
Description: RHO/H1FOO
Author: madelinebarlas
Session Name: mm9 RHO/H1FOO
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 281
|
1730966400 |
281 |
 |
Description: GB Multiomics custom data tracks - 2
Author: madisonflorenz
Session Name: GB Multiomics custom data tracks - 2
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 310
|
1730966400 |
310 |
 |
Description: mm9_NRL/WT
Author: l.g.altizer
Session Name: mm9_NRL/WT
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 283
|
1730966400 |
283 |
 |
Description: mm9 CRX CBR mRNA
Author: madelinebarlas
Session Name: mm9 CRX CBR mRNA
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 251
|
1730966400 |
251 |
 |
Description: 11/7/24
Author: aehodges
Session Name: mm9//2nd
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 288
|
1730966400 |
288 |
 |
Description: mm9 CBR regions
Author: ClaytonFannin
Session Name: mm9 CBR regions
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 276
|
1730966400 |
276 |
 |
Description: GB Multiomics custom data tracks - 1
Author: madisonflorenz
Session Name: GB Multiomics custom data tracks - 1
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 238
|
1730966400 |
238 |
 |
Description: 11/7/24
Author: aehodges
Session Name: mm9//1st
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 273
|
1730966400 |
273 |
 |
Description: mm9 CRX CBR
Author: madelinebarlas
Session Name: mm9 CRX CBR
Genome Assembly: mm9
Creation Date: 2024-11-07
Views: 285
|
1730966400 |
285 |
 |
Description: UCSC Genome browser features two custom tracks on chromosome 22, the regions from 20,100,000 to 20,140,000. The first track, titled as “spacer,” marks every 10,000 bases with blue ticks. The second track, named “even,” traced with red ticks every 100 bases (by skip 100 bases), with a 100-base interval between each tick. The “even” track shows labels such as : “first,” “second,” and “third.” These tracks illustrates how BED format can be used to visualize specific genomic intervals with determined position.
Author: ankathi.s
Session Name: Module8 assignment
Genome Assembly: hg38
Creation Date: 2024-11-07
Views: 273
|
1730966400 |
273 |
 |
Description: This UCSC Genome Browser session features two custom tracks on chromosome 22, covering the region from 20,100,000 to 20,140,000. The first track, titled “spacer,” marks every 10,000 bases with blue ticks. The second track, named “even,” uses red ticks every 100 bases, with a 100-base interval between each tick. The “even” track also includes labels for the first three ticks: “first,” “second,” and “third.” These tracks demonstrate how BED format can be used to visualize specific genomic intervals and annotations.
Author: das.arushi
Session Name: Module_8_assignment_hg38
Genome Assembly: hg38
Creation Date: 2024-11-06
Views: 298
|
1730880000 |
298 |
 |
Description: This session features two custom tracks on chromosome 22 on hg38 assembly, covering the region from 20,100,000 to 20,140,000. The first track, labeled "spacer" marks blue ticks every 10,000 bases. The second track, called "even" displays red ticks at intervals of 100 bases, with a 100-base gap between each tick. Additionally, the "even" track includes labels for the first three ticks: "first," "second," and "third." These custom tracks illustrate how BED format can be used to visualize specific genomic intervals and annotations.
Author: das.raj
Session Name: BINF 6310: Module 8 Assignment
Genome Assembly: hg38
Creation Date: 2024-11-06
Views: 271
|
1730880000 |
271 |
 |
Description: This session includes two custom tracks on chromosome 22, spanning the region 20100000-20140000. The first track, named 'spacer', displays blue ticks every 10,000 bases. The second track, named 'even', shows red ticks every 100 bases, skipping 100 bases between each tick. The 'even' track also includes labels for the first three ticks: 'first', 'second', and 'third'. These custom tracks demonstrate the use of BED format for visualizing specific genomic intervals and annotations.
Author: Pratham donda
Session Name: Custom Track Assignment
Genome Assembly: hg38
Creation Date: 2024-11-06
Views: 260
|
1730880000 |
260 |
 |
Description: This is ganesan murugan's session
Author: ganesan.m
Session Name: ganesan_murugan
Genome Assembly: hg38
Creation Date: 2024-11-06
Views: 282
|
1730880000 |
282 |
 |
Description: PDE6B:
macula RNA-seq
peripheral retina RNA-seq
Author: diazacex
Session Name: PDE6B
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 286
|
1730793600 |
286 |
 |
Description: RNA-seq custom tracks activity - PDE6B
Author: madisonflorenz
Session Name: RNA-seq custom tracks activity - PDE6B
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 301
|
1730793600 |
301 |
 |
Description: RNA-seq custom tracks activity - PDE6A
Author: madisonflorenz
Session Name: RNA-seq custom tracks activity - PDE6A
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 305
|
1730793600 |
305 |
 |
Description: PDE6A:
macula RNA-seq
peripheral retina RNA-seq
Author: diazacex
Session Name: PDE6A
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 293
|
1730793600 |
293 |
 |
Description: GNAT2:
macula RNA-seq
peripheral retina RNA-seq
Author: diazacex
Session Name: GNAT2_rec_mac
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 276
|
1730793600 |
276 |
 |
Description: GNAT 1:
macula RNA-seq
periperal retina RNA-seq
Author: diazacex
Session Name: GNAT1_ret_mac
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 320
|
1730793600 |
320 |
 |
Description: RNA-seq custom tracks activity - GNAT2
Author: madisonflorenz
Session Name: RNA-seq custom tracks activity - GNAT2
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 283
|
1730793600 |
283 |
 |
Description: RNA-seq custom tracks activity - GNAT1
Author: madisonflorenz
Session Name: RNA-seq custom tracks activity - GNAT1
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 279
|
1730793600 |
279 |
 |
Description: RHO macula RNA-seq
RHO periperal retina RNA-seq
Author: diazacex
Session Name: RHO_ret_mac
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 283
|
1730793600 |
283 |
 |
Description: 11/5
Author: aehodges
Session Name: hg38//custom tracks
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 232
|
1730793600 |
232 |
 |
Description: RNA-seq custom tracks activity - RHO
Author: madisonflorenz
Session Name: RNA-seq custom tracks activity - RHO
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 315
|
1730793600 |
315 |
 |
Description: Rhodopsin
Author: ALLDEREP
Session Name: hg38RHOMacula
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 276
|
1730793600 |
276 |
 |
Description: This custom track displays genomic intervals from the provided test.bed file, showing regions of interest for our class assignment. The data spans chromosomes 1 and 7 and highlights segments related to gene regulation and expression.
Author: Shamitha
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2024-11-05
Views: 325
|
1730793600 |
325 |
 |
Description: This session contains custom tracks visualizing specific features on chromosome 22 between positions 20,100,000 and 20,140,000. It includes:
Spacer Track:
Description: Blue ticks placed every 10,000 bases.
Color: Blue (RGB: 0, 0, 255)
Key Positions:
chr22:20100000-20100001
chr22:20110000-20110001
chr22:20120000-20120001
Even Track:
Description: Red ticks placed every 100 bases, with skips of 100 bases between ticks.
Color: Red (RGB: 255, 0, 0)
Key Positions:
chr22:20100000-20100100 (first)
chr22:20100200-20100300 (second)
chr22:20100400-20100500 (third)
Author: taliaroth19
Session Name: Module 8 Assignment
Genome Assembly: hg38
Creation Date: 2024-11-04
Views: 275
|
1730707200 |
275 |
 |
Description: customs tracks but better
Author: ALLDEREP
Session Name: hg38CustomTracksPractice - EA
Genome Assembly: hg38
Creation Date: 2024-10-31
Views: 268
|
1730361600 |
268 |
 |
Description: custom tracks
Author: ALLDEREP
Session Name: CustomTracks_Test
Genome Assembly: hg19
Creation Date: 2024-10-31
Views: 279
|
1730361600 |
279 |
 |
Description: CLIP2 is a gene that is related to linker protein function. Various RefSeq comparisons are aligned with expression levels from a variety of tissues.
Author: gwilds
Session Name: clip2
Genome Assembly: hg19
Creation Date: 2024-10-31
Views: 292
|
1730361600 |
292 |
 |
Description: MAPT mutations
Author: diazacex
Session Name: MAPT GB custom tracks
Genome Assembly: hg38
Creation Date: 2024-10-31
Views: 300
|
1730361600 |
300 |
 |
Description: MAPT mutations looged by Clayton Fannin
Author: ClaytonFannin
Session Name: hg38 MAPT mutations Group 5 (CAF)
Genome Assembly: hg38
Creation Date: 2024-10-31
Views: 292
|
1730361600 |
292 |
 |
Description: 10/31 Intro to UCSC Custom Data tracks
Author: madisonflorenz
Session Name: 10/31 Intro to UCSC Custom Data tracks
Genome Assembly: hg38
Creation Date: 2024-10-31
Views: 271
|
1730361600 |
271 |
 |
Description: 10/31/24
Author: aehodges
Session Name: hg38//MAPT gene
Genome Assembly: hg38
Creation Date: 2024-10-31
Views: 277
|
1730361600 |
277 |
 |
Description: UCSC GB custom tracks intro part 1A
Author: madisonflorenz
Session Name: UCSC GB custom tracks intro part 1A
Genome Assembly: hg19
Creation Date: 2024-10-31
Views: 299
|
1730361600 |
299 |
 |
Description: practicing custom tracks
Author: diazacex
Session Name: RHO Practice
Genome Assembly: hg19
Creation Date: 2024-10-31
Views: 278
|
1730361600 |
278 |
 |
Description: 10/31/24
Author: aehodges
Session Name: hg19//custom track
Genome Assembly: hg19
Creation Date: 2024-10-31
Views: 250
|
1730361600 |
250 |
 |
Description: ELK3 promoter region
Author: gauravagarwal
Session Name: ELK3_promoter
Genome Assembly: hg38
Creation Date: 2024-10-29
Views: 725
|
1730188800 |
725 |
 |
Description: Looking at the mutations in distal and tumor tissue of liver (hepatocellular carcinoma) in Patient #8. The mutations found exclusively in tumor tissue biopsy are highlighted in blue windows.
Author: dlk_browse
Session Name: Patient8_ND6
Genome Assembly: hg38
Creation Date: 2024-10-14
Views: 1173
|
1728892800 |
1173 |
 |
Description: Ridley Ch 4 HTT Fig 1
Author: madisonflorenz
Session Name: Ridley Ch 4 HTT Fig 1
Genome Assembly: hg38
Creation Date: 2024-10-14
Views: 439
|
1728892800 |
439 |
 |
Description: Ridley Ch 4 HTT Fig 2
Author: madisonflorenz
Session Name: Ridley Ch 4 HTT Fig 2
Genome Assembly: hg38
Creation Date: 2024-10-14
Views: 440
|
1728892800 |
440 |
 |
Description: 10/13/24
Author: aehodges
Session Name: hg38/HTT Gene
Genome Assembly: hg38
Creation Date: 2024-10-13
Views: 278
|
1728806400 |
278 |
 |
Description: THP-1 monocyte PMA timecourse
Author: Firefoot97
Session Name: T2T_monocyte_PMA_timecourse
Genome Assembly: hub_3671779_hs1
Creation Date: 2024-10-10
Views: 298
|
1728547200 |
298 |
 |
Description: Tracks H3K9me3 during E5 gametocytogenesis
Author: sandracula
Session Name: Sci-Rep_H3K9me3_gametocytes
Genome Assembly: hub_4042823_pfa2
Creation Date: 2024-10-04
Views: 453
|
1728028800 |
453 |
 |
Description: For Art & Katja
Author: mc.sierant
Session Name: RNU4-2_cons
Genome Assembly: hg38
Creation Date: 2024-09-30
Views: 663
|
1727683200 |
663 |
 |
Description: Genes e Microssatélites do Salminus brasiliensis set/2024 - LARGE_UFSJ
Author: jguimasr
Session Name: LARGE_UFSJ-GCA_030463535.1
Genome Assembly: hub_4414904_GCA_030463535.1
Creation Date: 2024-08-05
Views: 375
|
1722844800 |
375 |
 |
Description: Exon 10
Author: sofiaterenziani
Session Name: Exon 10 - HDG gene
Genome Assembly: hg38
Creation Date: 2024-09-26
Views: 268
|
1727337600 |
268 |
 |
Description: HDG_hg38
Author: l.g.altizer
Session Name: HDG_hg38
Genome Assembly: hg38
Creation Date: 2024-09-24
Views: 535
|
1727164800 |
535 |
 |
Description: HDG gene
Author: sofiaterenziani
Session Name: hg38 - HDG gene
Genome Assembly: hg38
Creation Date: 2024-09-24
Views: 466
|
1727164800 |
466 |
 |
Description: 9/24/24
Author: aehodges
Session Name: hg38//HGD session 2
Genome Assembly: hg38
Creation Date: 2024-09-24
Views: 463
|
1727164800 |
463 |
 |
Description: HGD gene - Figure 2
Author: madisonflorenz
Session Name: HGD gene - Figure 2
Genome Assembly: hg38
Creation Date: 2024-09-24
Views: 286
|
1727164800 |
286 |
 |
Description: Human genome HGD gene
Author: rchelsea
Session Name: hg38 HGD #1
Genome Assembly: hg38
Creation Date: 2024-09-24
Views: 246
|
1727164800 |
246 |
 |
Description: hg38HGDExon10
Author: ALLDEREP
Session Name: hg38HGDExon10
Genome Assembly: hg38
Creation Date: 2024-09-24
Views: 301
|
1727164800 |
301 |
 |
Description: HGD gene - Figure 1
Author: madisonflorenz
Session Name: HGD gene - Figure 1
Genome Assembly: hg38
Creation Date: 2024-09-24
Views: 312
|
1727164800 |
312 |
 |
Description: hg38 HGD
Author: ALLDEREP
Session Name: hg38 HGD
Genome Assembly: hg38
Creation Date: 2024-09-24
Views: 277
|
1727164800 |
277 |
 |
Description: edwcdewd
Author: maddiebendele
Session Name: HGD gene
Genome Assembly: hg38
Creation Date: 2024-09-23
Views: 297
|
1727078400 |
297 |
 |
Description: 9/20/24
Author: aehodges
Session Name: hg38//HGD gene
Genome Assembly: hg38
Creation Date: 2024-09-20
Views: 260
|
1726819200 |
260 |
 |
Description: Human chr2 compared to other mammals
Author: sofiaterenziani
Session Name: hg38 - chr2 + other organisms
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 494
|
1726560000 |
494 |
 |
Description: Human and other primates
Author: rchelsea
Session Name: hg38 chr2 #2
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 458
|
1726560000 |
458 |
 |
Description: 9/17/24
Author: aehodges
Session Name: hg38//session 2
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 487
|
1726560000 |
487 |
 |
Description: Region of Human Chr2 in Ridley Chapter and Other Primates
Author: madisonflorenz
Session Name: Region of Human Chr2 in Ridley Chapter and Other Primates
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 300
|
1726560000 |
300 |
 |
Description: wow
Author: maddiebendele
Session Name: Fusion_Event_Bendele
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 304
|
1726560000 |
304 |
 |
Description: Synteny and Conservation among Primates and the human
Author: ALLDEREP
Session Name: Synteny and Conservation among Primates
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 287
|
1726560000 |
287 |
 |
Description: Human and chimp chromosome #2
Author: rchelsea
Session Name: hg38 chr2
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 283
|
1726560000 |
283 |
 |
Description: hg38 chromosome 2 comparison
Author: ClaytonFannin
Session Name: hg38 chromosome 2 comparison
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 326
|
1726560000 |
326 |
 |
Description: region of human chromosome 2 discussed in the Ridley chapter
Author: madisonflorenz
Session Name: Region of Human Chr2 in Ridley Chapter
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 286
|
1726560000 |
286 |
 |
Description: Humans vs different Ape comparison of chr 3
Author: diazacex
Session Name: Comparison of Chr 3
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 285
|
1726560000 |
285 |
 |
Description: hg38 chromosome 2 full comparison
Author: ClaytonFannin
Session Name: hg38 chromosome 2 full comparison
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 315
|
1726560000 |
315 |
 |
Description: Human chromosome 2 in comparison to chimp chromosomes to show fusion.
Author: l.g.altizer
Session Name: Chimp_fusion
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 309
|
1726560000 |
309 |
 |
Description: Human fusion
Author: maddiebendele
Session Name: FusionEventBendele
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 305
|
1726560000 |
305 |
 |
Description: for prez
Author: maddiebendele
Session Name: p.aergiona presentation
Genome Assembly: hub_3560215_GCF_000006765.1
Creation Date: 2024-09-17
Views: 325
|
1726560000 |
325 |
 |
Description: Ridley Ch2 Species Pt. 1
Author: madisonflorenz
Session Name: Ridley Ch2 Species Pt. 1
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 340
|
1726560000 |
340 |
 |
Description: Comparison on Chr2 with chimps
Author: sofiaterenziani
Session Name: hg38 - chr2
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 282
|
1726560000 |
282 |
 |
Description: human chr2 synteny to chimps
Author: marte2je
Session Name: hg38 chimp - chr2
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 279
|
1726560000 |
279 |
 |
Description: comparison of the KPNA6 gene from humans to opossums
Author: ALLDEREP
Session Name: M.Domestica Comparison
Genome Assembly: hg19
Creation Date: 2024-09-15
Views: 292
|
1726387200 |
292 |
 |
Description: H3K27ac ChIP-seq and RNA-seq of lung neuroendocrine tumors from GSE247574.
Author: yotamd
Session Name: lung_NET
Genome Assembly: hg38
Creation Date: 2024-09-10
Views: 673
|
1725955200 |
673 |
 |
Description: CRX gene isoforms
Author: madisonflorenz
Session Name: CRX gene isoforms
Genome Assembly: hg38
Creation Date: 2024-09-10
Views: 521
|
1725955200 |
521 |
 |
Description: CRX gene
Author: ClaytonFannin
Session Name: CRX gene BIO481 CAF
Genome Assembly: hg38
Creation Date: 2024-09-10
Views: 548
|
1725955200 |
548 |
 |
Description: 9/10
Author: aehodges
Session Name: CRX Gene
Genome Assembly: hg38
Creation Date: 2024-09-10
Views: 305
|
1725955200 |
305 |
 |
Description: RHO
Author: ALLDEREP
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 328
|
1725264000 |
328 |
 |
Description: The custom tracts represent Hop1 ChIP-Seq data in wild type, ATPase deficient mutant and null mutant. Bam files contains uniquely mapped reads to S. cerevisiae genome. The sample information is given below:
1. m_sorted_mapped_cer_HP1_4h_Ch_1.bam- Hop1 WT replicate 1,
2. m_sorted_mapped_cer_HP1_4h_Ch_2.bam- Hop1 WT replicate 2,
3. m_sorted_mapped_cer_HP1_4h_In_1.bam- Hop1 WT input,
4. m_sorted_mapped_cer_SK_Hp1m_4h_Ch1.bam- Hop1 ATPase mutant replicate 1,
5. m_sorted_mapped_cer_SK_Hp1m_4h_Ch2.bam- Hop1 ATPase mutant replicate 2,
6. m_sorted_mapped_cer_SK_Hp1m_4h_In2.bam- Hop1 ATPase mutant input,
7. m_sorted_mapped_cer_hp1_4h_Ch_1.bam- Hop1 null mutant repliacte 1,
8. m_sorted_mapped_cer_hp1_4h_Ch_2.bam- Hop1 null mutant replicate 2,
9. m_sorted_mapped_cer_hp1_4h_In_1.bam- Hop1 null mutant input
Author: Sameerjoshi1997
Session Name: Hop1 ChIP-Seq data
Genome Assembly: sacCer3
Creation Date: 2024-09-04
Views: 653
|
1725436800 |
653 |
 |
Description: Quiz 2
Author: sofiaterenziani
Session Name: hg19 RHO 2
Genome Assembly: hg19
Creation Date: 2024-09-03
Views: 525
|
1725350400 |
525 |
 |
Description: Abigail Hodges
Author: aehodges
Session Name: hg19 RHO redo
Genome Assembly: hg19
Creation Date: 2024-09-03
Views: 534
|
1725350400 |
534 |
 |
Description: Quiz 1
Author: sofiaterenziani
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-09-03
Views: 314
|
1725350400 |
314 |
 |
Description: RHO
Author: diazacex
Session Name: RHO_hg19
Genome Assembly: hg19
Creation Date: 2024-09-03
Views: 312
|
1725350400 |
312 |
 |
Description: hg19 RHO
Author: madelinebarlas
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-09-03
Views: 308
|
1725350400 |
308 |
 |
Description: RHO gene and OMIM Alleles
Author: l.g.altizer
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-09-03
Views: 300
|
1725350400 |
300 |
 |
Description: RHO Gene & OMIM & Multi
Author: l.g.altizer
Session Name: hg19_RHO_2
Genome Assembly: hg38
Creation Date: 2024-09-03
Views: 301
|
1725350400 |
301 |
 |
Description: hg19 RHO - quiz #2
Author: madisonflorenz
Session Name: hg19 RHO - quiz #2
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 254
|
1725264000 |
254 |
 |
Description: hg19 RHO - Quiz 1
Author: gplaughon912
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 282
|
1725264000 |
282 |
 |
Description: hg19 RHO - Quiz 2
Author: gplaughon912
Session Name: hg19 RHO - Session 2
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 331
|
1725264000 |
331 |
 |
Description: RHO NEW
Author: ALLDEREP
Session Name: hg19RHO NEW
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 279
|
1725264000 |
279 |
 |
Description: same as the previous rho but with highlight
Author: maddiebendele
Session Name: hg19 RHO2
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 283
|
1725264000 |
283 |
 |
Description: Quiz 2
Author: aehodges
Session Name: hg19 RHO session 2
Genome Assembly: hg19
Creation Date: 2024-09-01
Views: 293
|
1725177600 |
293 |
 |
Description: hg19 RHO
Author: ClaytonFannin
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-09-01
Views: 289
|
1725177600 |
289 |
 |
Description: It is tracked by samples and treatment groups. Some differentially accessible peaks highlighted
Author: Pengxin Yang
Session Name: pig_BCG_ATAC_filtered_by_sample_and_merged_highlight
Genome Assembly: susScr11
Creation Date: 2025-03-03
Views: 181
|
1740988800 |
181 |
 |
Description: Fall24 MouseBIO481 group5 (CAF)
Author: ClaytonFannin
Session Name: Fall24 MouseBIO481 group5 (CAF)
Genome Assembly: mm9
Creation Date: 2024-08-29
Views: 310
|
1724918400 |
310 |
 |
Description: Fall24 Worm BIO481 group#3
Author: madisonflorenz
Session Name: Fall24 Worm BIO481 group#3
Genome Assembly: ce11
Creation Date: 2024-08-29
Views: 247
|
1724918400 |
247 |
 |
Description: First experience using UCSC Genome Browser looking at the Rhodopsin gene of a mouse
Author: ALLDEREP
Session Name: Fall24 MouseBIO481 group5
Genome Assembly: mm9
Creation Date: 2024-08-29
Views: 308
|
1724918400 |
308 |
 |
Description: C.elegans ICD-1
Author: marte2je
Session Name: Fall24 Worm Bio481 group 3 #2
Genome Assembly: ce11
Creation Date: 2024-08-29
Views: 237
|
1724918400 |
237 |
 |
Description: Fall24 MouseBIO481 group6
Author: madelinebarlas
Session Name: Fall24 MouseBIO481 group6
Genome Assembly: mm9
Creation Date: 2024-08-29
Views: 314
|
1724918400 |
314 |
 |
Description: CGEMS Mouse AEH Test
Author: aehodges
Session Name: Fall24 MouseBIO481 group6
Genome Assembly: mm9
Creation Date: 2024-08-29
Views: 324
|
1724918400 |
324 |
 |
Description: Fall24 Worm BIO481 group#3
C. elegans
Author: sofiaterenziani
Session Name: Fall24 Worm BIO481 group#3
Genome Assembly: ce11
Creation Date: 2024-08-29
Views: 273
|
1724918400 |
273 |
 |
Description: Fall24 Worm BIO481 group 3
Author: rchelsea
Session Name: Fall24 Worm BIO481 group 3
Genome Assembly: ce11
Creation Date: 2024-08-29
Views: 348
|
1724918400 |
348 |
 |
Description: MEK
Author: diazacex
Session Name: Fall24 Yeast BIO481 group#2
Genome Assembly: sacCer3
Creation Date: 2024-08-29
Views: 248
|
1724918400 |
248 |
 |
Description: In class assignment 8/29
Author: Mitcheij
Session Name: Fall24 Worm BIO481 group 4
Genome Assembly: ce11
Creation Date: 2024-08-29
Views: 307
|
1724918400 |
307 |
 |
Description: C. elegans ICD-1
Author: marte2je
Session Name: Fall24 Worm Bio481 group 3
Genome Assembly: ce11
Creation Date: 2024-08-29
Views: 289
|
1724918400 |
289 |
 |
Description: BIO481 UCSC Genome Browser Assignment - Group 5 Mouse
Author: gplaughon912
Session Name: Fall24 MouseBIO481 Group5
Genome Assembly: mm9
Creation Date: 2024-08-29
Views: 317
|
1724918400 |
317 |
 |
Description: CRX
Author: ALLDEREP
Session Name: hg38 CRX
Genome Assembly: hg38
Creation Date: 2024-09-12
Views: 302
|
1726128000 |
302 |
 |
Description: For Bioinformatics class assignment purpose, to be viewed by the instructors.
Author: hemib
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2024-10-08
Views: 312
|
1728374400 |
312 |
 |
Description: CUT&RUN dataset for Six3 (with antibodies Six3_abs and Six3_ori), Six6, and IgG control for mouse embryonic retinas at E13.5.
Author: Wei_einstein
Session Name: PlosOne
Genome Assembly: mm10
Creation Date: 2024-08-08
Views: 1199
|
1723104000 |
1199 |
 |
Description: hg19 RHO altered for quiz 2
Author: ClaytonFannin
Session Name: hg19 RHO altered
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 280
|
1725264000 |
280 |
 |
Description: GWAS endometriosis associations around RERG
Author: Pkgg77
Session Name: RERG - ras ER reg high in uterus CHR12
Genome Assembly: hg19
Creation Date: 2024-07-22
Views: 1729
|
1721635200 |
1729 |
 |
Description: MPAC predictions on all of Gnomad. See html files associated with tracks and https://www.biorxiv.org/content/10.1101/2025.04.16.648420v1.full for more information
Author: saarantras
Session Name: MPAC_gnomad
Genome Assembly: hg38
Creation Date: 2025-06-09
Views: 92
|
1749456000 |
92 |
 |
Description: These datasets are part of the manuscript title: Interpreting regulatory mechanisms of Hippo signaling through a deep learning sequence model.
https://doi.org/10.1101/2024.02.22.580842
Example browser position : chr15:102015496-102017061
Author: KhyatiD
Session Name: Dalal_hippo_pathway_TFs_mm10
Genome Assembly: mm10
Creation Date: 2025-02-19
Views: 226
|
1739952000 |
226 |
 |
Description: This session shows how UShER can place H5N1 sequences on the tree of viral samples. Here are the steps to replicate a similar session. First find an example viral sequence such as SRR28752449_HA_cns.fa in this repository: https://github.com/andersen-lab/avian-influenza/tree/master/fasta
With the fasta sequence copied follow these steps.
1. Navigate to UShER, quick link: https://usher.bio
2. Paste in a sequence:
>Consensus_SRR28752449_HA_cns_threshold_0.5_quality_20
ATGGAGAACATAG...
3. Click "Upload"
4. Click "View in Genome Browser"
5. Note the placement near the branch of H5N1 near other cattle lineages (click session image on the left to see, find the light blue horizontal line highlighting Consensus_SRR28752449).
Author: brianlee
Session Name: H5N1
Genome Assembly: hub_4086371_GCF_000864105.1
Creation Date: 2024-07-01
Views: 1726
|
1719820800 |
1726 |
 |
Description: Chromatin accessibility tracks for human B cell multiome dataset.
Author: peasena321
Session Name: Human_Bcell_Multiome_Tracks
Genome Assembly: hg38
Creation Date: 2025-03-18
Views: 236
|
1742284800 |
236 |
 |
Description: hg38 CNGB1 alt polyA isoforms
Author: renkejhsph
Session Name: hg38 CNGB1 alt polyA isoforms
Genome Assembly: hg38
Creation Date: 2024-06-19
Views: 1262
|
1718784000 |
1262 |
 |
Description: Archaic introgression identified with the use of hmmix by Skov et al., (https://github.com/LauritsSkov/Introgression-detection). Genotype data from 1000 genomes projects.
Author: daxa213
Session Name: hg19_SkovHMM_1
Genome Assembly: hg19
Creation Date: 2024-10-29
Views: 300
|
1730188800 |
300 |
 |
Description: Trackplots Including WT, Chx10-Cre;Ptbp1lox/+(Het1-3) and Chx10-Cre;Ptbp1lox/lox(KO), together with trackplots from ASCOT (Ling et al 2020)
Author: roggercarmen
Session Name: Ptbp1KO_Retina
Genome Assembly: mm10
Creation Date: 2025-07-01
Views: 88
|
1751356800 |
88 |
 |
Description: HumanDevelopingRetinaAtlas
Author: zhenzuo2
Session Name: HumanDevelopingRetinaAtlas
Genome Assembly: hg38
Creation Date: 2024-06-06
Views: 1163
|
1717660800 |
1163 |
 |
Description: "A two-step regulatory mechanism dynamically controls histone H3 acetylation by SAGA complex at growth-related promoters" paper genome wide data (Zencir et al., 2024)
Author: sevil
Session Name: GSE268170_BIGWIG FILES
Genome Assembly: sacCer3
Creation Date: 2024-06-05
Views: 737
|
1717574400 |
737 |
 |
Description: All QTLs from datasets in our S. cerevisiae QTL datasets reanalysis.
QTL Track Name Format:
- GENE::dataset_name$$LOD, where LOD is rounded to the nearest integer
Relevant Fields:
- Color: blue for mRNA QTLs, brown for protein QTLs, black for xQTLs
- Color Intensity: darker with increasing QTL LOD
- Strand: direction of QTL as considered in the paper
- Arrows: right for positive QTL effect
Author: matthewf
Session Name: Reanalysis of QTL Datasets
Genome Assembly: sacCer3
Creation Date: 2024-08-05
Views: 335
|
1722844800 |
335 |
 |
Description: long retinal isoform of shisa5
Author: JonathanM70
Session Name: hg38 shisa5 long isoform
Genome Assembly: hg38
Creation Date: 2024-06-03
Views: 598
|
1717401600 |
598 |
 |
Description: Chip-seq
Author: ucscfzl
Session Name: SIRT3KO
Genome Assembly: hg38
Creation Date: 2024-08-21
Views: 345
|
1724227200 |
345 |
 |
Description: nair
Author: xiaopei
Session Name: hg38_xiaopei_May20
Genome Assembly: hg38
Creation Date: 2024-05-20
Views: 819
|
1716192000 |
819 |
 |
Description: test
Author: xiaopei
Session Name: hg38_test2
Genome Assembly: hg38
Creation Date: 2024-05-20
Views: 725
|
1716192000 |
725 |
 |
Description: test 1
Author: xiaopei
Session Name: hg38_test1
Genome Assembly: hg38
Creation Date: 2024-05-20
Views: 600
|
1716192000 |
600 |
 |
Description: ATAC-seq YZ
Author: Yichao Zhou
Session Name: NewATAC-seq analysis
Genome Assembly: mm10
Creation Date: 2024-05-20
Views: 310
|
1716192000 |
310 |
 |
Description: hg18_rnaseq
Author: hyfydky
Session Name: hg18_rnaseq
Genome Assembly: hg18
Creation Date: 2024-05-18
Views: 397
|
1716019200 |
397 |
 |
Description: hg38_GSE176016
Author: hyfydky
Session Name: hg38_GSE176016
Genome Assembly: hg38
Creation Date: 2024-05-10
Views: 993
|
1715328000 |
993 |
 |
Description: c.4619T>G P.Leu1540Arg Revel score
Author: sk202
Session Name: VWF_1540
Genome Assembly: hg19
Creation Date: 2024-05-16
Views: 408
|
1715846400 |
408 |
 |
Description: test
Author: yunna
Session Name: test
Genome Assembly: hg38
Creation Date: 2024-05-02
Views: 613
|
1714636800 |
613 |
 |
Description: isPCR hits in two adjacent genes. Shows amplicons expected if using mRNA as template.
Author: netherlands2024
Session Name: hg19_isPCR
Genome Assembly: hg19
Creation Date: 2024-04-29
Views: 1489
|
1714377600 |
1489 |
 |
Description: Microsatellites Density Landscape (Main track)
Including sub-tracks: Landscape of STR Density, HMDP, MMDP, LMDP
Description: "The microsatellite, also called short tandem repeats (STRs), density landscapes illustrated the relative density of local microsatellites per kilobase of sequence along the complete reference Human Genome T2T-CHM13, indicating that microsatellites tend to aggregate in a characteristic manner, resulting in numerous microsatellite density peaks throughout the genome. These microsatellite density peaks can be categorized into three distinct types: High Microsatellite Density Peak (HMDP), Middle Microsatellite Density Peak (MMDP), and Low Microsatellite Density Peak (LMDP). Therefore, the main track contains four sub-tracks which are Landscape of STR Density, HMDP, MMDP, LMDP.
Methods: Each chromosome sequence of T2T-CHM13 was divided into a large number of units (bins) by the differential method with a resolution of 1kb. The local microsatellite relative density in every 1kb resolution unit was referred as position-related Differential-unit1kb (D1) Relative Density (pD1RD) and calculated by formula: pD1RDi = mi/1kb × 1000.
Track statistics summary:
Genome Assembled Size (bp): 3,117,275,501
Total peak count: 244,729
HMDP count: 8,823
MMDP count: 36,257
LMDP count: 199,649
Microsatellites Density Landscape
References:
Xia Y, Li D, Chen T, Pan S, Huang H, Zhang W, Liang Y, Fu Y, Peng Z, Zhang H et al. Microsatellite density landscapes illustrate short tandem repeats aggregation in the complete reference human genome. BMC Genomics. 2024; 25(1):960. PMID: 39402450; DOI: 10.1186/s12864-024-10843-9
Li D, Pan S, Zhang H, Fu Y, Peng Z, Zhang L, et al. A comprehensive microsatellite landscape of human Y-DNA at kilobase resolution. BMC genomics. 2021; 22(1):76. PMID: 33482734; PMCID: PMC7821415
Author: zhongyangtan
Session Name: CHM13v2.0
Genome Assembly: hub_3671779_hs1
Creation Date: 2023-09-10
Views: 769
|
1694332800 |
769 |
 |
Description: CRX binding sites for HTT gene in mouse genome
Author: eraterman
Session Name: mm9 HTT
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 1796
|
1712736000 |
1796 |
 |
Description: CRX binding sites for THRB gene in mouse genome
Author: eraterman
Session Name: mm9 THRB
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 1413
|
1712736000 |
1413 |
 |
Description: CRX binding sites for PDE6A gene in mouse genome
Author: eraterman
Session Name: mm9 PDE6A
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 694
|
1712736000 |
694 |
 |
Description: CRX binding sites for RCVRN gene in mouse genome
Author: eraterman
Session Name: mm9 RCVRN
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 439
|
1712736000 |
439 |
 |
Description: CRX binding sites for RHO gene in mouse genome
Author: eraterman
Session Name: mm9 RHO
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 428
|
1712736000 |
428 |
 |
Description: class assignment 4-10-24
Author: ShMaBe
Session Name: CRX ChIP Seq wt Nrl
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 406
|
1712736000 |
406 |
 |
Description: lab 4-3
Author: Ruffi2ec
Session Name: Lab due 4-3
Genome Assembly: hg38
Creation Date: 2024-04-04
Views: 412
|
1712217600 |
412 |
 |
Description: This session contains a custom track showing near coding regions in PanelApp green genes, as described in the article "Systematic identification of disease-causing promoter and untranslated region variants in 8,040 undiagnosed individuals with rare disease" Martin-Geary et al 2024
Author: ageary
Session Name: CRDG 'Near coding regions' Martin-Geary et al '24
Genome Assembly: hg38
Creation Date: 2024-04-05
Views: 469
|
1712304000 |
469 |
 |
Description: yes
Author: aryanzandi123
Session Name: REST and Co-repressor Window
Genome Assembly: hg38
Creation Date: 2024-12-11
Views: 315
|
1733904000 |
315 |
 |
Description: a demo
Author: xiyuan09
Session Name: xyuan2024-hg19
Genome Assembly: hg19
Creation Date: 2024-03-30
Views: 649
|
1711785600 |
649 |
 |
Description: U2AF2 iCLIP (Sutandy et al 2018) and eCLIP (ENCODE) processed with racoon_clip
Author: melinak
Session Name: U2AF2_iCLIP_and_eCLIP
Genome Assembly: hg38
Creation Date: 2024-04-12
Views: 441
|
1712908800 |
441 |
 |
Description: Ribosome Footprinting profile of miR181a/b-1 knockout vs wildtype in mouse thymocytes with three samples each. Data from Verheyden and Klostermann et al. 2024 (https://www.biorxiv.org/content/10.1101/2023.09.08.556730v1)
Author: melinak
Session Name: miR181_RibosomeFootprint_Verheyden&Klostermann_2024
Genome Assembly: mm10
Creation Date: 2024-03-26
Views: 805
|
1711440000 |
805 |
 |
Description: hg19 RHO #2 Quiz #@
Author: rchelsea
Session Name: hg19 RHO #2
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 232
|
1725264000 |
232 |
 |
Description: hg19 RHO Quiz #1
Author: rchelsea
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 299
|
1725264000 |
299 |
 |
Description: hg19 RHO
Author: madisonflorenz
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2024-09-02
Views: 305
|
1725264000 |
305 |
 |
Description: miR181-eCLIP data from Verheyden and Klostermann et.al. 2024. Shown are the chimeric and non-chimeric crosslinks for four conditions:
Ago2 IP
Ago2 IP with enrichment for miR181
Ago2 IP with miR181a/b-1 knockout
Ago2 IP with miR181a/b-1 knockout and a following enrichment for miR181
In addition binding sites are shown for
Ago2 IP
Ago2 IP with enrichment for miR181
Ago2 IP with miR181a/b-1 knockout
And the differential binding sites of Ago2 IP with miR181a/b-1 knockout vs Ago2 IP
Author: melinak
Session Name: miR181-eCLIP_Verheyden&Klostermann_2024
Genome Assembly: mm10
Creation Date: 2024-03-25
Views: 563
|
1711353600 |
563 |
 |
Description: This browser track accompanies the Alternative Proteome Detection Project out of the Sheynkman Lab at the University of Virginia.
Author: emilyfwatts
Session Name: Alternative-Proteome-Detection
Genome Assembly: hg38
Creation Date: 2024-03-15
Views: 610
|
1710489600 |
610 |
 |
Description: Rearrangements of the 17q24.3 region associated with abnormal phenotypes.
Author: 429035671
Session Name: Pei_et_al_2024_fig6
Genome Assembly: hg19
Creation Date: 2024-03-11
Views: 957
|
1710144000 |
957 |
 |
Description: Test session for Module 7 of BINF 6310.
Author: oconnor.ea
Session Name: oconnor-binf6310
Genome Assembly: hg38
Creation Date: 2024-03-01
Views: 1113
|
1709280000 |
1113 |
 |
Description: Blue ticks every 10000 bases. Red ticks every 100 bases, skip 100
Author: lizkiefer
Session Name: hg38_chr22_KieferLiz
Genome Assembly: hg38
Creation Date: 2024-02-27
Views: 1586
|
1709020800 |
1586 |
 |
Description: ATAC-seq of MaSC(mammary stem cell)
Author: woaichixiangcai
Session Name: MaSC_ATAC
Genome Assembly: mm10
Creation Date: 2024-02-18
Views: 908
|
1708243200 |
908 |
 |
Description: Track Decorators help page:
https://genome.ucsc.edu/goldenPath/help/decorator.html
Author: BGMP2024
Session Name: Track_Decorators
Genome Assembly: hg38
Creation Date: 2024-01-28
Views: 882
|
1706428800 |
882 |
 |
Description: Highlighted exons 5-8 of ENST00000269305.9
Author: BGMP2024
Session Name: hg38_BGMP2024_Ex
Genome Assembly: hg38
Creation Date: 2024-01-28
Views: 1002
|
1706428800 |
1002 |
 |
Description: practice
Author: jordancibellis
Session Name: hg19_480L
Genome Assembly: hg19
Creation Date: 2024-01-24
Views: 817
|
1706083200 |
817 |
 |
Description: BIO480L Spring 24
Author: penguinttoes
Session Name: hg19_newsome
Genome Assembly: hg19
Creation Date: 2024-01-24
Views: 565
|
1706083200 |
565 |
 |
Description: This is Harshini and Jenn's steps to solve FAD!
Author: creminslab
Session Name: FAD_Harshini_hg38
Genome Assembly: hg38
Creation Date: 2024-01-19
Views: 643
|
1705651200 |
643 |
 |
Description: te
Author: aryanzandi123
Session Name: Polycomb and Chromatin-Binding Proteins Window
Genome Assembly: hg38
Creation Date: 2024-12-11
Views: 257
|
1733904000 |
257 |
 |
Description: This session features a track hub with MaveDB experiment datasets mapped to the GRCh38 reference genome
Author: mcline
Session Name: MaveDB
Genome Assembly: hg38
Creation Date: 2024-04-03
Views: 592
|
1712131200 |
592 |
 |
Description: PE2 editing efficiency in synHEK3 reporters in K562 cells
Author: lxyttpp912
Session Name: hg38_synHEK3_hub
Genome Assembly: hg38
Creation Date: 2023-12-26
Views: 1068
|
1703577600 |
1068 |
 |
Description: Differentially methylated CGIs under DiPRO/ZNF555 knockdown in human rhabdomyosarcoma and myoblast cells (shDiPRO1 vs. non-targeting shCTL)
Author: dr_finder
Session Name: metCGI_ZNF55/DiPRO1_2023
Genome Assembly: hg38
Creation Date: 2023-12-20
Views: 431
|
1703059200 |
431 |
 |
Description: Coverage tracks from SF3B1-humanized yeast treated with splicing inhibitors Plad-B and Thail-A from Hunter et al. Published in RNA
Author: mannyares
Session Name: Hunter_etal
Genome Assembly: sacCer3
Creation Date: 2023-12-13
Views: 1000
|
1702454400 |
1000 |
 |
Description: may
Author: xiaopei
Session Name: hg38_xiaopei_May21
Genome Assembly: hg38
Creation Date: 2024-05-21
Views: 380
|
1716278400 |
380 |
 |
Description: bigWig files for promoter NDR resetting after replication (Zencir et al.,2024)
Author: julien_soudet
Session Name: EdU_replication
Genome Assembly: sacCer3
Creation Date: 2024-05-31
Views: 394
|
1717142400 |
394 |
 |
Description: CTCF loops, Oxford nanopore phased methylation and RNAseq in LUHMES cell line for the 15q11-q13 region. View of AS imprinted region as shown in Figure 7A of Gutierrez Fugon et al, Human Molecular Genetics, 2024.
Author: ojg333
Session Name: AS_Locus_Fig7A_Fugon_2024
Genome Assembly: hg19
Creation Date: 2023-12-06
Views: 398
|
1701849600 |
398 |
 |
Description: A snapshot from the genome browser displaying the enrichment of H3K27ac in the replicates of the two groups, with and without sodium lactate.Yanhua Zhao, Meiting Zhang, Xingwei Huang, Jiqiang Liu, Yuchen Sun, Fan Zhang, Na Zhang and Lei Lei Lactate modulates zygotic genome activation though H3K18 lactylation rather than H3K27 acetylation.
Author: ZhangMeiTing
Session Name: H3K27ac
Genome Assembly: mm10
Creation Date: 2023-12-03
Views: 809
|
1701590400 |
809 |
 |
Description: The UCSC Browser show the enrichment of the H3K27ac signal of Eif2b5 in both +L and -L groups.Yanhua Zhao, Meiting Zhang, Xingwei Huang, Jiqiang Liu, Yuchen Sun, Fan Zhang, Na Zhang and Lei Lei Lactate modulates zygotic genome activation though H3K18 lactylation rather than H3K27 acetylation.
Author: ZhangMeiTing
Session Name: Eif2b5_H3K27ac
Genome Assembly: mm10
Creation Date: 2023-12-03
Views: 624
|
1701590400 |
624 |
 |
Description: The UCSC Browser show the enrichment of the H3K27ac signal of Ccnt1 in both +L and -L groups.Yanhua Zhao, Meiting Zhang, Xingwei Huang, Jiqiang Liu, Yuchen Sun, Fan Zhang, Na Zhang and Lei Lei Lactate modulates zygotic genome activation though H3K18 lactylation rather than H3K27 acetylation.
Author: ZhangMeiTing
Session Name: Ccnt1_H3K27ac
Genome Assembly: mm10
Creation Date: 2023-12-03
Views: 588
|
1701590400 |
588 |
 |
Description: The UCSC Browser show the enrichment of the H3K27ac signal of Dppa2 in both +L and -L groups.Yanhua Zhao, Meiting Zhang, Xingwei Huang, Jiqiang Liu, Yuchen Sun, Fan Zhang, Na Zhang and Lei Lei Lactate modulates zygotic genome activation though H3K18 lactylation rather than H3K27 acetylation.
Author: ZhangMeiTing
Session Name: Dppa2_H3K27ac
Genome Assembly: mm10
Creation Date: 2023-12-03
Views: 520
|
1701590400 |
520 |
 |
Description: The UCSC Browser show the enrichment of the H3K18la signal of Ccnt1 in both +L and -L groups.Yanhua Zhao, Meiting Zhang, Xingwei Huang, Jiqiang Liu, Yuchen Sun, Fan Zhang, Na Zhang and Lei Lei Lactate modulates zygotic genome activation though H3K18 lactylation rather than H3K27 acetylation.
Author: ZhangMeiTing
Session Name: Ccnt1_H3K18la
Genome Assembly: mm10
Creation Date: 2023-12-03
Views: 514
|
1701590400 |
514 |
 |
Description: The UCSC Browser show the enrichment of the H3K18la signal of Eif2b5 in both +L and -L groups.Yanhua Zhao, Meiting Zhang, Xingwei Huang, Jiqiang Liu, Yuchen Sun, Fan Zhang, Na Zhang and Lei Lei Lactate modulates zygotic genome activation though H3K18 lactylation rather than H3K27 acetylation.
Author: ZhangMeiTing
Session Name: Eif2b5_H3K18la
Genome Assembly: mm10
Creation Date: 2023-12-03
Views: 522
|
1701590400 |
522 |
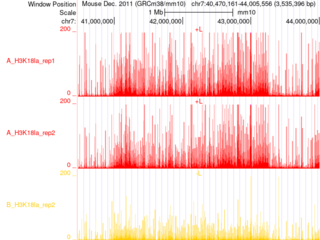 |
Description: A snapshot from the genome browser displaying the enrichment of H3K18la in the replicates of the two groups, with and without sodium lactate.Yanhua Zhao, Meiting Zhang, Xingwei Huang, Jiqiang Liu, Yuchen Sun, Fan Zhang, Na Zhang and Lei Lei Lactate modulates zygotic genome activation though H3K18 lactylation rather than H3K27 acetylation.
Author: ZhangMeiTing
Session Name: H3K18la
Genome Assembly: mm10
Creation Date: 2023-12-03
Views: 530
|
1701590400 |
530 |
 |
Description: The UCSC Browser show the enrichment of the H3K18la signal of Dppa2 in both +L and -L groups.Yanhua Zhao, Meiting Zhang, Xingwei Huang, Jiqiang Liu, Yuchen Sun, Fan Zhang, Na Zhang and Lei Lei Lactate modulates zygotic genome activation though H3K18 lactylation rather than H3K27 acetylation.
Author: ZhangMeiTing
Session Name: Dppa2_H3K18la
Genome Assembly: mm10
Creation Date: 2023-12-03
Views: 522
|
1701590400 |
522 |
 |
Description: ZC3 ChIP
Author: yannfrey
Session Name: ZC3 ChIP
Genome Assembly: hg38
Creation Date: 2023-11-26
Views: 674
|
1700985600 |
674 |
 |
Description: CTCF loops, Oxford nanopore phased methylation and RNAseq in LUHMES cell line for the 15q11-q13 region. Zoomed in view of the MAGEL2 anchor region as shown in Figure 9A of Gutierrez Fugon et al, Human Molecular Genetics, 2024.
Author: ojg333
Session Name: MAGEL2_Fig9A _Fugon_2024
Genome Assembly: hg19
Creation Date: 2024-07-22
Views: 339
|
1721635200 |
339 |
 |
Description: RNA omics stuff
Author: nrpowell
Session Name: 2023_Nov_CYP3A4_fold/m6a/eclip_comparison
Genome Assembly: hg38
Creation Date: 2023-11-20
Views: 817
|
1700467200 |
817 |
 |
Description: Tdg&p53
Author: shaozhenyu2017@sibcb.ac.cn
Session Name: Tdg&p53
Genome Assembly: mm10
Creation Date: 2023-11-18
Views: 513
|
1700294400 |
513 |
 |
Description: PDE6C&RBP4
Author: gracewattawa
Session Name: PDE6C&RBP4
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 542
|
1699948800 |
542 |
 |
Description: p
Author: julianna0220
Session Name: PDE6C
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 606
|
1699948800 |
606 |
 |
Description: GUCA1B&GUCA1A
Author: gracewattawa
Session Name: GUCA1B&GUCA1A
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 418
|
1699948800 |
418 |
 |
Description: d
Author: julianna0220
Session Name: GUCA1B
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 552
|
1699948800 |
552 |
 |
Description: mm9 PDE6C/RBP4 Multiomics
Author: chelseadonovan
Session Name: mm9 PDE6C/RBP4 Multiomics
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 539
|
1699948800 |
539 |
 |
Description: NRL&CPNE6
Author: gracewattawa
Session Name: NRL&CPNE6
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 393
|
1699948800 |
393 |
 |
Description: d
Author: julianna0220
Session Name: cone6
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 566
|
1699948800 |
566 |
 |
Description: nrl version of everything
Author: joshraynes
Session Name: Nrl version
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 542
|
1699948800 |
542 |
 |
Description: RHO&H1foo
Author: gracewattawa
Session Name: RHO&H1foo rnaseq
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 363
|
1699948800 |
363 |
 |
Description: pho
Author: julianna0220
Session Name: RHO Pho
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 538
|
1699948800 |
538 |
 |
Description: mm9 GUCA1B/GUCA1A Multiomics
Author: chelseadonovan
Session Name: mm9 GUCA1B/GUCA1A Multiomics
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 504
|
1699948800 |
504 |
 |
Description: mm9 NRL/CPNE6 Multiomics
Author: chelseadonovan
Session Name: mm9 NRL/CPNE6 Multiomics
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 539
|
1699948800 |
539 |
 |
Description: RNASEQnrl
Author: gracewattawa
Session Name: RNASEQnrl
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 529
|
1699948800 |
529 |
 |
Description: mm9 RHO Multiomics
Author: chelseadonovan
Session Name: mm9 RHO Multiomics
Genome Assembly: mm9
Creation Date: 2023-11-14
Views: 546
|
1699948800 |
546 |
 |
Description: BINF6310
Author: varun1010
Session Name: hg38_assignment7_1
Genome Assembly: hg38
Creation Date: 2023-11-12
Views: 568
|
1699776000 |
568 |
 |
Description: RCVRN gene
Author: Isabella Lindblad
Session Name: 11-9_mouse3_RCVRN
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 556
|
1699516800 |
556 |
 |
Description: mm9 RCVRN CBR sites
Author: chelseadonovan
Session Name: mm9 RCVRN CBR sites
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 537
|
1699516800 |
537 |
 |
Description: RCVRN
Author: kenzie.prettyman
Session Name: RCVRN
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 544
|
1699516800 |
544 |
 |
Description: rcvrn
Author: julianna0220
Session Name: RCVRN
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 535
|
1699516800 |
535 |
 |
Description: HTT
Author: kenzie.prettyman
Session Name: HTT
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 604
|
1699516800 |
604 |
 |
Description: mm9 HTT CBR sites
Author: chelseadonovan
Session Name: mm9 HTT CBR sites
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 531
|
1699516800 |
531 |
 |
Description: HTT
Author: julianna0220
Session Name: HTT
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 560
|
1699516800 |
560 |
 |
Description: CRX binding regions of PDE6A
Author: taylo2ga
Session Name: Mouse PDE6A
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 535
|
1699516800 |
535 |
 |
Description: THRB
Author: kenzie.prettyman
Session Name: THRB
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 573
|
1699516800 |
573 |
 |
Description: thrb
Author: julianna0220
Session Name: THRB
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 552
|
1699516800 |
552 |
 |
Description: mm9 THRB CBR sites
Author: chelseadonovan
Session Name: mm9 THRB CBR sites
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 565
|
1699516800 |
565 |
 |
Description: PDE6A
Author: kenzie.prettyman
Session Name: PDE6A
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 551
|
1699516800 |
551 |
 |
Description: RHO nrl- onlh
Author: joshraynes
Session Name: RHO Nrl-
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 509
|
1699516800 |
509 |
 |
Description: PDE6A
Author: julianna0220
Session Name: PDE6A
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 552
|
1699516800 |
552 |
 |
Description: mm9 PDE6A CBR sites
Author: chelseadonovan
Session Name: mm9 PDE6A CBR sites
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 546
|
1699516800 |
546 |
 |
Description: Viewing Mouse Genome 2007 (mm9), RHO gene with CRX binding sites
Author: chelseadonovan
Session Name: mm9 RHO CBR sites
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 563
|
1699516800 |
563 |
 |
Description: crx binding
Author: julianna0220
Session Name: CRX JP
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 542
|
1699516800 |
542 |
 |
Description: CBR data in PDE6A gene on mouse mm9 genome
Author: ryleestrother
Session Name: CBR PDE6A mm9
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 529
|
1699516800 |
529 |
 |
Description: CBR regions of the RCVRN gene
Author: taylo2ga
Session Name: Mouse RCVRN
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 533
|
1699516800 |
533 |
 |
Description: crx binding sites
Author: gracewattawa
Session Name: CRX binding sites
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 546
|
1699516800 |
546 |
 |
Description: CRX binding regions of the gene HTT
Author: taylo2ga
Session Name: Mouse HTT
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 533
|
1699516800 |
533 |
 |
Description: CRX binding sites wt and mutant
Author: kenzie.prettyman
Session Name: CRX
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 594
|
1699516800 |
594 |
 |
Description: Enke Bio481 THRB Gene in mm9
Author: cooperki
Session Name: THRB_Mouse
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 554
|
1699516800 |
554 |
 |
Description: RHO gene
Author: Isabella Lindblad
Session Name: 11-9_mouse_RHO
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 523
|
1699516800 |
523 |
 |
Description: CRX binding regions of the THRB gene
Author: taylo2ga
Session Name: Mouse THRB
Genome Assembly: mm9
Creation Date: 2023-11-09
Views: 515
|
1699516800 |
515 |
 |
Description: binf6310 hg38 custom track
Author: sydneycole
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-11-05
Views: 686
|
1699171200 |
686 |
 |
Description: test.bed custom tracks
Author: rpatel01
Session Name: binfmodule7
Genome Assembly: hg38
Creation Date: 2023-11-03
Views: 937
|
1698998400 |
937 |
 |
Description: BINF6310 MODULE 7
Author: Pruthweish27
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 843
|
1698739200 |
843 |
 |
Description: BINF6310
MODULE 7
Author: sreepoojitha
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-11-02
Views: 562
|
1698912000 |
562 |
 |
Description: ancestry - taster nontaster
Author: fareehaa_ahm
Session Name: ancestry
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 553
|
1698912000 |
553 |
 |
Description: hg19 TAS2R38 Ancestry Data
Author: chelseadonovan
Session Name: hg19 TAS2R38 Ancestry Data
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 550
|
1698912000 |
550 |
 |
Description: ancestry
Author: julianna0220
Session Name: Ancestry.com
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 540
|
1698912000 |
540 |
 |
Description: Ancestry.com SNPs
Author: ryleestrother
Session Name: hg19 ancestry.com
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 499
|
1698912000 |
499 |
 |
Description: Ancestry.com data
Author: kenzie.prettyman
Session Name: Ancestry.com
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 548
|
1698912000 |
548 |
 |
Description: custom tracks - BIO 481
Author: fareehaa_ahm
Session Name: MYBPC1_customtracks_Fareeha
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 574
|
1698912000 |
574 |
 |
Description: MYBPC1
Author: julianna0220
Session Name: MYBPC1 JP
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 521
|
1698912000 |
521 |
 |
Description: Final with descriptions
Author: rahrigHK
Session Name: MYBPC1 Activity (Final)
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 530
|
1698912000 |
530 |
 |
Description: MYBPC1 session
Author: joshraynes
Session Name: MYBPC1
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 543
|
1698912000 |
543 |
 |
Description: MYBPC1 assignment
Author: kenzie.prettyman
Session Name: MYBPC1
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 563
|
1698912000 |
563 |
 |
Description: hg19 assembly showing MYBPC1 gene & custom tracks
Author: chelseadonovan
Session Name: hg19 MYBPC1
Genome Assembly: hg19
Creation Date: 2023-11-02
Views: 537
|
1698912000 |
537 |
 |
Description: BINF_6310 Module 7
Author: kandalkarankita
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-11-02
Views: 542
|
1698912000 |
542 |
 |
Description: BINF-6310
Author: Harshini144
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-11-02
Views: 531
|
1698912000 |
531 |
 |
Description: BINF 6310
Module 7
Author: nallaboluvikas
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-11-01
Views: 1201
|
1698825600 |
1201 |
 |
Description: Custom tracks for genomic variants on chr22
Author: sdzzwzy
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2023-11-01
Views: 542
|
1698825600 |
542 |
 |
Description: BINF6310
Author: SaradaPriya_Mns
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 774
|
1698739200 |
774 |
 |
Description: binf6310
Author: varun1010
Session Name: hg38_assignment7
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 652
|
1698739200 |
652 |
 |
Description: BINF 6310
Author: Pavitra Dass
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 548
|
1698739200 |
548 |
 |
Description: BINF6310
Author: Meghana09
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 639
|
1698739200 |
639 |
 |
Description: custom link for assignment7
Author: srivastava.sank
Session Name: assignment7track
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 539
|
1698739200 |
539 |
 |
Description: DAG session
Author: DaniGonzalez
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2024-10-31
Views: 298
|
1730361600 |
298 |
 |
Description: Incorporating a customized track
Author: Tanvi
Session Name: test
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 579
|
1698739200 |
579 |
 |
Description: BINF6310 Assignment: Incorporating a customized track using the provided bed file into the UCSC Genome Browser.
Submitted by AashkaB
Author: aashka.b
Session Name: test_session_AB
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 560
|
1698739200 |
560 |
 |
Description: This is a session run as a part of assignment for the course BINF6310
Author: Unnati Jonnavithula
Session Name: UJ1
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 530
|
1698739200 |
530 |
 |
Description: this is the assignment
Author: yuqiaotang
Session Name: 6300assignment7
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 580
|
1698739200 |
580 |
 |
Description: Module 7 - BIF6310
Author: trangtu97
Session Name: hg38_human
Genome Assembly: hg38
Creation Date: 2023-10-30
Views: 564
|
1698652800 |
564 |
 |
Description: test for school
Author: tojaga.m
Session Name: Eosinophil
Genome Assembly: hg38
Creation Date: 2023-10-26
Views: 554
|
1698307200 |
554 |
 |
Description: Trout
Author: Rafet
Session Name: ChromHmm
Genome Assembly: hub_2243217_GCF_013265735.2
Creation Date: 2023-10-25
Views: 43
|
1698220800 |
43 |
 |
Description: Harvard MCB197 course, histone modifications in human tissues
Author: amandajoyward
Session Name: MCB197-histone
Genome Assembly: hg38
Creation Date: 2023-10-20
Views: 547
|
1697788800 |
547 |
 |
Description: hg38 Blat of in situ hybridization probes to NTRK2.
Author: amandajoyward
Session Name: MCB197-NTRK2 probes
Genome Assembly: hg38
Creation Date: 2023-10-30
Views: 526
|
1698652800 |
526 |
 |
Description: BINF 6310
Author: pavan kumar
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-10-30
Views: 526
|
1698652800 |
526 |
 |
Description: Quiz 10/12
Author: jschifflin
Session Name: HTT gene assignment
Genome Assembly: hg38
Creation Date: 2023-10-12
Views: 855
|
1697097600 |
855 |
 |
Description: hg38 HTT gene BIO481 - 10/12/23
Author: fareehaa_ahm
Session Name: hg38_HTT
Genome Assembly: hg38
Creation Date: 2023-10-12
Views: 718
|
1697097600 |
718 |
 |
Description: hg38 genome viewing HTT gene with GENCODE V32 and Simple Repeats
Author: chelseadonovan
Session Name: hg38 HTT gene
Genome Assembly: hg38
Creation Date: 2023-10-11
Views: 1080
|
1697011200 |
1080 |
 |
Description: htt gene
Author: julianna0220
Session Name: hg38 HTT gene
Genome Assembly: hg38
Creation Date: 2023-10-11
Views: 601
|
1697011200 |
601 |
 |
Description: haethethntgnarangr
Author: jrkenne
Session Name: hg19_hbg
Genome Assembly: hg19
Creation Date: 2023-10-02
Views: 859
|
1696233600 |
859 |
 |
Description: Hemoglobin
Author: Biniyam
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2023-09-28
Views: 700
|
1695888000 |
700 |
 |
Description: The genes HBB and HBD are both expressed in red blood cells, but HBB is also expressed in many other tissues, whereas HBD in expressed in red blood cells almost exclusively.
Author: laparidis.e
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2023-11-05
Views: 543
|
1699171200 |
543 |
 |
Description: Methylation data for human and mouse
Author: masarun
Session Name: masa_test
Genome Assembly: hg38
Creation Date: 2023-09-21
Views: 1128
|
1695283200 |
1128 |
 |
Description: BAF and histone markers in SS mouse model
Author: Li Li
Session Name: mouse_SS_BAF_histoneMarkers
Genome Assembly: mm10
Creation Date: 2024-06-27
Views: 390
|
1719475200 |
390 |
 |
Description: Chip seq
Author: xiaopei
Session Name: Chip_seq_A
Genome Assembly: hg38
Creation Date: 2024-06-27
Views: 366
|
1719475200 |
366 |
 |
Description: Microsatellites Density Landscape (Main track)
Including sub-tracks: Landscape of STR Density, HMDP, MMDP, LMDP
Description: The landscapes illustrated the relative density of local microsatellites per kilobase of sequence along the comprehensive reference Human Genome GRCh38 , indicating that microsatellites tend to aggregate in a characteristic manner, resulting in numerous peaks of microsatellite density throughout the genome. These microsatellite density peaks can be categorized into three distinct types: High Microsatellite Density Peak (HMDP), Middle Microsatellite Density Peak (MMDP), and Low Microsatellite Density Peak (LMDP). Therefore, the main track contains four sub-tracks which are Landscape of STR Density, HMDP, MMDP, LMDP.
Methods: Each chromosome sequence of GRCh38 was divided into a large number of units (bins) by the differential method with a resolution of 1kb. The local microsatellite relative density in every 1kb resolution unit was referred as position-related Differential-unit1kb (D1) Relative Density (pD1RD) and calculated by formula: pD1RDi = mi/1kb × 1000.
Track statistics summary:
Genome Assembled Size (bp): 3,088,269,832
Total peak count: 208,638
HMDP count: 3,480
MMDP count: 26,310
LMDP count: 178,848
Session Name: Microsatellite Density Landscape
References:
Xia Y, Li D, Chen T, Pan S, Huang H, Zhang W, Liang Y, Fu Y, Peng Z, Zhang H et al. Microsatellite density landscapes illustrate short tandem repeats aggregation in the complete reference human genome. BMC Genomics. 2024; 25(1):960. PMID: 39402450; DOI: 10.1186/s12864-024-10843-9
Li D, Pan S, Zhang H, Fu Y, Peng Z, Zhang L, et al. A comprehensive microsatellite landscape of human Y-DNA at kilobase resolution. BMC genomics. 2021; 22(1):76. PMID: 33482734; PMCID: PMC7821415
Author: zhongyangtan
Session Name: GRCh38.p14
Genome Assembly: hg38
Creation Date: 2023-09-10
Views: 560
|
1694332800 |
560 |
 |
Description: zoom in PBK and find out all the upstream 200base SNPs
Author: ZhengJi
Session Name: hg38_upstream200SNPs
Genome Assembly: hg38
Creation Date: 2023-09-05
Views: 1229
|
1693900800 |
1229 |
 |
Description: probes for FISH on human cells and tissues
Author: rafal.czapiewski@ed.ac.uk
Session Name: hg19_FISH_probes
Genome Assembly: hg19
Creation Date: 2024-01-17
Views: 541
|
1705478400 |
541 |
 |
Description: Crouch DRIP-Seq rDNA annotations
Author: apratim.mitra
Session Name: crouch-dripseq-rdna-annotations-diff
Genome Assembly: hub_4200124_mm10
Creation Date: 2023-08-31
Views: 251
|
1693468800 |
251 |
 |
Description: class 3
Author: hekn
Session Name: “Fall23 Worm BIO481 group 3
Genome Assembly: ce11
Creation Date: 2023-08-31
Views: 630
|
1693468800 |
630 |
 |
Description: Beginner Activity for UCSC Genome Browser (BIO481)
Author: fareehaa_ahm
Session Name: Fall23 Worm BIO481 group4
Genome Assembly: ce11
Creation Date: 2023-08-31
Views: 662
|
1693468800 |
662 |
 |
Description: genomic 481 in class activity
Author: ryleestrother
Session Name: Fall23 Yeast BIO481 group2
Genome Assembly: sacCer3
Creation Date: 2023-08-31
Views: 576
|
1693468800 |
576 |
 |
Description: Group Activity #1 from Group 1.
Author: jschifflin
Session Name: Fall23 Yeast BIO481 Group1
Genome Assembly: sacCer3
Creation Date: 2023-08-31
Views: 593
|
1693468800 |
593 |
 |
Description: group 5 version of mouse project
Author: julianna0220
Session Name: Fall23 Mouse481 group5
Genome Assembly: mm9
Creation Date: 2023-08-31
Views: 564
|
1693468800 |
564 |
 |
Description: Mouse genome group project group 6
Author: joshraynes
Session Name: Fall23 MouseBio481 Group 6
Genome Assembly: mm9
Creation Date: 2023-08-31
Views: 527
|
1693468800 |
527 |
 |
Description: Mouse genome (mm9) for Group #6 in BIO481
Author: chelseadonovan
Session Name: Fall23 Mouse BIO481 Genome Group#6
Genome Assembly: mm9
Creation Date: 2023-08-31
Views: 390
|
1693468800 |
390 |
 |
Description: The genes HBB and HBD are both expressed in red blood cells, but HBB is also expressed in many other tissues, whereas HBD is expressed in red cells almost exclusively.
Author: shancon1200
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2023-08-31
Views: 569
|
1693468800 |
569 |
 |
Description: Show every Lysine site on POMC 1st extron on hg38.
Author: ZhengJi
Session Name: hg38_POMC1stextronK
Genome Assembly: hg38
Creation Date: 2023-08-31
Views: 585
|
1693468800 |
585 |
 |
Description: hg18_rnaseq3
Author: hyfydky
Session Name: hg18_rnaseq3
Genome Assembly: hg18
Creation Date: 2024-05-20
Views: 406
|
1716192000 |
406 |
 |
Description: BTD MANE in HG38 UCSC browser
Author: lilinatera
Session Name: hg38_BTD_MANE
Genome Assembly: hg38
Creation Date: 2023-08-27
Views: 712
|
1693123200 |
712 |
 |
Description: Clinically relevant pesudogenes sequences [high homology: orange; low homology: light orange, as per Mandelker et al. 2016; PMID: 27228465. Example: ALG1_0.26_241_1_241; Gene Name _ Mappability _ Positions <1 _ % Positions <1 _ Max contiguous bases; the 'ALG1' gene feature at position chr16:5127843-5128083, Exon+/-65bp has a 'Mappabilty score' of 0.26 (1= mapping only once to the genome whereas a score lower than 1 equates to homology else where on the genome. 241 'Positions' (bases) affected, 100'% of these Positions' and 241 'Max contiguous bases' within the Exon+/-65bp that are affected as indicated by having a mappability score below 1.
Author: Yog77
Session Name: hg19_ClinicalHomology
Genome Assembly: hg19
Creation Date: 2017-01-17
Views: 1842
|
1484640000 |
1842 |
 |
Description: Archaic introgression identified with the use of hmmix by Skov et al., (https://github.com/LauritsSkov/Introgression-detection). Genotype data from 1000 genomes projects.
Author: daxa213
Session Name: hg19_SkovHMM_GBR_CHS
Genome Assembly: hg19
Creation Date: 2023-08-23
Views: 281
|
1692777600 |
281 |
 |
Description: 2
Author: jschifflin
Session Name: hg19 RHO 2
Genome Assembly: hg19
Creation Date: 2023-09-05
Views: 583
|
1693900800 |
583 |
 |
Description: This session is a view of the 10 capstone genes in the SSPsyGene project. It uses the multi-region tool to slice the genome over several chromosomes to display all the genes in one view. Capstone genes:ARID1B ASXL3 CACNA1G CHD8 DLL1 GABRA1 KMT2C SCN2A SHANK3 SMARCC2
Author: brianlee
Session Name: Capstone_Genes
Genome Assembly: hg38
Creation Date: 2023-08-15
Views: 1335
|
1692086400 |
1335 |
 |
Description: This session is a view of the 10 capstone genes in the SSPsyGene project in order of chromosomes (a gene on chr2 before the gene on chr5). It uses the multi-region tool to slice the genome over several chromosomes to display all the genes in one view. Capstone genes:SCN2A GABRA1 DLL1 ARID1B KMT2C SMARCC2 CHD8 CACNA1G ASXL3 SHANK3
Author: brianlee
Session Name: CapstoneGenes
Genome Assembly: hg38
Creation Date: 2023-08-15
Views: 725
|
1692086400 |
725 |
 |
Description: Testing public session
Author: gperez2
Session Name: Testing_public_session
Genome Assembly: hg38
Creation Date: 2023-08-03
Views: 666
|
1691049600 |
666 |
 |
Description: BioInf 6310 bed file upload
Author: Neveen Mohamed
Session Name: nv23
Genome Assembly: hg38
Creation Date: 2023-10-31
Views: 563
|
1698739200 |
563 |
 |
Description: ATAC-seq pile-up tracks for fruit fly clock neurons.
For more details and citation: https://doi.org/10.1101/2023.08.15.553315
Author: yegeyy
Session Name: ATAC wildtype and per01 pile-up tracks
Genome Assembly: dm6
Creation Date: 2023-08-08
Views: 599
|
1691481600 |
599 |
 |
Description: Test
Author: gperez2
Session Name: brca1
Genome Assembly: hg38
Creation Date: 2023-10-27
Views: 553
|
1698393600 |
553 |
 |
Description: E5ind and NF54-GEXP02-Tom H3K9me3 dynamics at the onset of gam. dev.
Author: sandracula
Session Name: H3K9me3 dynamics at the onset of gam. dev.
Genome Assembly: hub_2790993_GCA_000002765.3
Creation Date: 2024-03-12
Views: 463
|
1710230400 |
463 |
 |
Description: d
Author: julianna0220
Session Name: hg19 RHO JP
Genome Assembly: hg19
Creation Date: 2023-09-04
Views: 558
|
1693814400 |
558 |
 |
Description: eclip comparison 2023
Author: nrpowell
Session Name: eCLIP_comparison_May_2023
Genome Assembly: hg38
Creation Date: 2023-05-23
Views: 2565
|
1684828800 |
2565 |
 |
Description: This custom session includes manually selected blood-related DNA methylation data measured by bisulfite sequencing. This initial session include 121 tracks updated until 2023 June.
Author: dai905
Session Name: hg38_bloodcell_121
Genome Assembly: hg38
Creation Date: 2023-06-02
Views: 545
|
1685692800 |
545 |
 |
Description: rs142783062
Author: nrpowell
Session Name: rs142783062
Genome Assembly: hg38
Creation Date: 2023-06-01
Views: 2186
|
1685606400 |
2186 |
 |
Description: Epigenetic selectivity of TopoIIα redistribution and histone eviction of anthracyclines
Anthracyclines act by disrupting the interface of TopoII and DNA and by evicting histones. The anthracycline-specific redistribution of TopoII and its association with histone eviction were addressed in K562 cells. Chromatin immunoprecipitation, followed by deep sequencing (ChIP-seq) against endogenously tagged TopoIIα, and transposase-accessible chromatin with sequencing (ATAC-seq) was performed 4 hours after anthracycline exposure.
Author: mtan1
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2023-08-11
Views: 628
|
1691740800 |
628 |
 |
Description: Data Viewer
Author: allenbellamom
Session Name: hg38_DC
Genome Assembly: hg38
Creation Date: 2023-10-25
Views: 544
|
1698220800 |
544 |
 |
Description: GR ChIP
Author: tsuzuki
Session Name: PC_hg38
Genome Assembly: hg38
Creation Date: 2023-05-17
Views: 2336
|
1684310400 |
2336 |
 |
Description: This session describes the genomic location for LD-SNPs of rs7255249 (r2>0.8). (Top) The plot shows the eQTL association of local variants with CHST8 expression in breast mammary tissues. (Middle) GeneHancer track shows probable interactions between regulatory elements and CHST8 promoter. (Bottom) DHS region, H3K4me1, H3K4me3, and H3K27ac marks present putative epigenetic regulation.
DHS signals of each cells are from ENCODE. Histone modification signals of MCF7, HMEC-1 (mammary epithelial cell, 50-year-old female), and HMEC-2 (breast epithelium, 51-year-old female) are also from ENCODE, whereas histone modification results of vHMEC (Breast_vHMEC.Donor_RM035) and BMC (Breast_Myoepithelial_Cells) are from Roadmap Epigenomics.
Histone modification signals from different biosamples are grouped in order by H3K4me3, H3K4me1, and H3K27ac and presented them as overlaid tracks.
Author: Wen-Cheng
Session Name: Breast_rs7255249
Genome Assembly: hg19
Creation Date: 2023-05-18
Views: 562
|
1684396800 |
562 |
 |
Description: UCSC browser session associated with manuscript:
PMID: 38032818
Title: Chromosome-specific maturation of the epigenome in the Drosophila male germline
Author: jamesanderson12358
Session Name: analysis230508___UCSC_session_germline_MS
Genome Assembly: dm6
Creation Date: 2023-05-08
Views: 696
|
1683532800 |
696 |
 |
Description: inputA.bed and inputA.bedgraph
Author: amykle
Session Name: inputA from usr 16
Genome Assembly: hg38
Creation Date: 2023-05-04
Views: 848
|
1683187200 |
848 |
 |
Description: hugo example RTS
Author: Example
Session Name: hg38_hugo2023
Genome Assembly: hg38
Creation Date: 2023-05-04
Views: 804
|
1683187200 |
804 |
 |
Description: Mouse meth
Author: danilapror
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2023-05-04
Views: 888
|
1683187200 |
888 |
 |
Description: OMICS_HW_2_Sokolov_AA
Author: hazirliver1
Session Name: mm10_Sokolov_AA
Genome Assembly: mm10
Creation Date: 2023-05-03
Views: 794
|
1683100800 |
794 |
 |
Description: WGBS and RNA-seq data from ENCODE
- T cells
- GN12878
- NK
- Pancr
end_OTHERS
Looking at gene CD55!
Author: michalula
Session Name: WGBS_RNA_ENCODE_T_GN12878_NK_Pancr_end_OTHERS_CD55
Genome Assembly: hg38
Creation Date: 2023-05-01
Views: 1117
|
1682928000 |
1117 |
 |
Description: dnasel
Author: aehodges
Session Name: mm9/3.4.25
Genome Assembly: mm9
Creation Date: 2025-03-04
Views: 184
|
1741075200 |
184 |
 |
Description: PSIP ChIP (GSE175482)
Author: tsuzuki
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-04-09
Views: 1332
|
1681027200 |
1332 |
 |
Description: Gene project for genetics class
Author: bji20
Session Name: Gene Project Final
Genome Assembly: hg38
Creation Date: 2023-04-07
Views: 1050
|
1680854400 |
1050 |
 |
Description: Tracks from GSE212386. Titration of H3K9me3 and H3K27ac antibodies in ChIP, as well as replicates of H3K9me2 IPs.
Author: ari11
Session Name: GSE212386
Genome Assembly: hg38
Creation Date: 2023-04-05
Views: 995
|
1680681600 |
995 |
 |
Description: CUT&RUN data for "Stress increases sperm respiration and motility in mice and men."
Numbers in the sample names are associated with treatment condition as follows:
Vehicle: 4, 5, 10, 12
Cort: 2, 3, 7, 8, 9
Author: nickole.kanyuch
Session Name: CUT&RUN_NM_03-2023
Genome Assembly: mm10
Creation Date: 2023-03-28
Views: 241
|
1679990400 |
241 |
 |
Description: BS assay and gRNA tracks for ATRT synthetic lethal candidate genes
Author: matejboros
Session Name: hg19_custom_track
Genome Assembly: hg19
Creation Date: 2023-03-07
Views: 1236
|
1678176000 |
1236 |
 |
Description: This is the session showing the location and expression of the new junction that might cause frame shift.
Author: dandan_0909
Session Name: hg38_5_26
Genome Assembly: hg38
Creation Date: 2025-05-26
Views: 259
|
1748246400 |
259 |
 |
Description: Tissue-level Atlas of Mouse Isoforms.
We used full-length cDNA sequencing and the Mandalorion tool to identify and quantify high confidence transcript Isoforms for a variety of mouse tissues
Author: vollmers
Session Name: TAMI
Genome Assembly: mm39
Creation Date: 2024-01-25
Views: 885
|
1706169600 |
885 |
 |
Description: gene_expression_GM12878
Author: rafal.czapiewski@ed.ac.uk
Session Name: hg19_expression_GM12878
Genome Assembly: hg19
Creation Date: 2023-01-24
Views: 1429
|
1674547200 |
1429 |
 |
Description: these are the 4 variants to show bruce CTs in sessions
Author: cincinnati2023
Session Name: hg38_4vars
Genome Assembly: hg38
Creation Date: 2023-01-26
Views: 822
|
1674720000 |
822 |
 |
Description: Wyatt Dano Bioinformatics hands on session gene public session
Author: wpd20
Session Name: MCR1 Gene
Genome Assembly: hg38
Creation Date: 2023-10-23
Views: 537
|
1698048000 |
537 |
 |
Description: Human Chromosome 22 from the Human Genome Consortium build 38 (the latest annotated reference assembly of the human genome)
Author: elittlestone
Session Name: HGC/38
Genome Assembly: hg38
Creation Date: 2023-10-23
Views: 533
|
1698048000 |
533 |
 |
Description: tRNA
Author: ck-j
Session Name: tRNA
Genome Assembly: hg38
Creation Date: 2022-12-16
Views: 825
|
1671177600 |
825 |
 |
Description: hg19 RHO gene with Multiz Allignments and Basewise Conservation (phyloP)
Author: chelseadonovan
Session Name: hg19 RHO Multiz Alignments
Genome Assembly: hg19
Creation Date: 2023-09-05
Views: 571
|
1693900800 |
571 |
 |
Description: ChIP-seq of Histone 3 Acetylation (H3ac) marks.
Author: HanZhang-
Session Name: H3ac_ChIP
Genome Assembly: hg38
Creation Date: 2023-08-11
Views: 513
|
1691740800 |
513 |
 |
Description: This data has been made public to provide access to peer reviewers
Author: VetrieLab
Session Name: BV173 ChIP Datasets_Scott et al
Genome Assembly: hg38
Creation Date: 2022-11-15
Views: 752
|
1668499200 |
752 |
 |
Description: BALB/c mouse tissue level transcriptome atlas
Author: msadams
Session Name: BALB/c mouse transcriptome
Genome Assembly: mm39
Creation Date: 2022-08-09
Views: 835
|
1660032000 |
835 |
 |
Description: Contribution tracks for peak 2
Author: lacey.walker
Session Name: peak2
Genome Assembly: hg38
Creation Date: 2022-10-15
Views: 2870
|
1665820800 |
2870 |
 |
Description: Contribution tracks for peak 0
Author: lacey.walker
Session Name: peak0
Genome Assembly: hg38
Creation Date: 2022-10-15
Views: 1791
|
1665820800 |
1791 |
 |
Description: Contribution tracks for peak 1
Author: lacey.walker
Session Name: peak1
Genome Assembly: hg38
Creation Date: 2022-10-15
Views: 876
|
1665820800 |
876 |
 |
Description: Genome-wide characterization of mitochondrial DNA methylation in human brain.
Matthew Devall1, Darren M. Soanes, Adam R. Smith, Emma L. Dempster, Rebecca G. Smith, Joe Burrage, Artemis Iatrou, Eilis Hannon, Claire Troakes, Karen Moore, Paul O’Neill, Safa Al-Sarraj, Leonard Schalkwyk, Jonathan Mill, Michael Weedon & Katie Lunnon
The mitochondrial genome is characterized by specific regions of methylation. Graph showing average % methylation at each cytosine in the mitochondrial genome from superior temporal gyrus (STG) and cerebellum (CER) brain tissue from seven donors free of neurodegenerative disease, including three males and four females using a targeted bisulfite sequencing method.
Author: dmsoanes
Session Name: chrM_methylation_STG_CER
Genome Assembly: hg38
Creation Date: 2022-10-04
Views: 992
|
1664870400 |
992 |
 |
Description: Used for CMSE 411
Author: CVan0124
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2022-09-23
Views: 794
|
1663920000 |
794 |
 |
Description: HIV-1 strain NL4-3 mRNA Atlas v1 mRNA transcript models open reading frames (ORFs) mapped onto HIV-1 RefSeq:NC_001802.1.
Author: Alejandro Gener. Contact: itspronouncedhenner@gmail.com
Transcript model method: Transcript models were downloaded as FASTA from GenBank:MZ242719.1-MZ242757.1 were blat (sic) against NC_001802.1 (GCF_000864765.1) in UCSC. Max number of sequences for blat = 25, so n=38 was partitioned into two blat runs, sorted by accession. These were viewed as a track in UCSC Genome Browser. MZ242719.1, MZ242720.1, MZ242721.1 are models of unspliced, singly-spliced, and doubly-spliced. "Singly-spliced" and "doubly-spliced" have been commonly used in the literature, but without basis in explicitly defined mRNA models. As you can see, HIV-1 NL4-3 splice isoforms supported by high-quality long reads and reanalysis are more complicated that previously understood.
Open reading frames (ORFs) method: Coding features were downloaded as FASTA from GenBank:MZ242719.1 were blat (sic) against NC_001802.1 (GCF_000864765.1) in UCSC. These were viewed as a track in UCSC Genome Browser.
ORF note 1: Exact ranges for GENER10 and GENER12 differ because of apparent sequence differences at their start codons. This is because blat did not match the small leading exon 5'-ATGGCAG-3' which spans NC_001802.1:4599-4605. This exon is supported by long-read mRNA of NL4-3 infected and transfected cells.
ORF note 2: Small regions of homology overlap polypurine tracts important for viral replication. These are not ORFs.
ORF note 3: MZ242719.1 (NL4-3) and NC_001802.1 are both HIV-1 subgroup B viruses. NL4-3 is a synthetic chimeric virus reviewed elsewhere. However, it is the most common strain of HIV used in laboratory settings. Both are modeled as "R-U5-HIV-U3-R".
If using the NL4-3 mRNA Atlas v1, please cite:
1.) Gener, A. R. (2022). Anticipating HIV drug resistance with appropriate sequencing methods. AIDS, 36(1). https://journals.lww.com/aidsonline/Fulltext/2022/01010/Anticipating_HIV_drug_resistance_with_appropriate.16.aspx.
2.) Gener, A. R., Klotman, P. E., Danesh, F. R., Cijiang He, J., Kimata, J. T., Lupski, J. R., & Ross, M. J. (2021). HIV informatics . Baylor College of Medicine Integrative Molecular and Biomedical Sciences Graduate Program.
Author: gener
Session Name: GCF_000864765.1_HIV-1_mRNA_Atlas_v1_blat_mRNA+ORFs
Genome Assembly: hub_3521609_GCF_000864765.1
Creation Date: 2022-09-21
Views: 596
|
1663747200 |
596 |
 |
Description: HIV-1 strain NL4-3 mRNA Atlas v1 open reading frames (ORFs) mapped onto HIV-1 RefSeq:NC_001802.1.
Author: Alejandro Gener. Contact: itspronouncedhenner@gmail.com
Method: Coding features were downloaded as FASTA from GenBank:MZ242719.1 were blat (sic) against NC_001802.1 (GCF_000864765.1) in UCSC. These were viewed as a track in UCSC Genome Browser.
Note 1: Exact ranges for GENER10 and GENER12 differ because of apparent sequence differences at their start codons. This is because blat did not match the small leading exon 5'-ATGGCAG-3' which spans NC_001802.1:4599-4605. This exon is supported by long-read mRNA of NL4-3 infected and transfected cells.
Note 2: Small regions of homology overlap polypurine tracts important for viral replication. These are not ORFs.
Note 3: MZ242719.1 (NL4-3) and NC_001802.1 are both HIV-1 subgroup B viruses. NL4-3 is a synthetic chimeric virus reviewed elsewhere. However, it is the most common strain of HIV used in laboratory settings. Both are modeled as "R-U5-HIV-U3-R".
If using the NL4-3 mRNA Atlas v1, please cite:
1.) Gener, A. R. (2022). Anticipating HIV drug resistance with appropriate sequencing methods. AIDS, 36(1). https://journals.lww.com/aidsonline/Fulltext/2022/01010/Anticipating_HIV_drug_resistance_with_appropriate.16.aspx.
2.) Gener, A. R., Klotman, P. E., Danesh, F. R., Cijiang He, J., Kimata, J. T., Lupski, J. R., & Ross, M. J. (2021). HIV informatics . Baylor College of Medicine Integrative Molecular and Biomedical Sciences Graduate Program.
Author: gener
Session Name: GCF_000864765.1_HIV-1_mRNA_Atlas_v1_blat_ORFs
Genome Assembly: hub_3521609_GCF_000864765.1
Creation Date: 2022-09-20
Views: 1065
|
1663660800 |
1065 |
 |
Description: This session is used for CNV interpretation
Author: JZhaoARUP
Session Name: hg38_ARUP_CMA_multi_gene
Genome Assembly: hg38
Creation Date: 2022-09-19
Views: 1316
|
1663574400 |
1316 |
 |
Description: This session is used for CNV interpretation
Author: JZhaoARUP
Session Name: hg38_ARUP_CMA_single_gene
Genome Assembly: hg38
Creation Date: 2022-09-19
Views: 807
|
1663574400 |
807 |
 |
Description: eCLIP experiments with ligation step allowing identification of miRNA-mRNA pairs and binding sites to around 60 nt resolution. Data generated in the laboratory of Todd Skaar.
Author: nrpowell
Session Name: miRNA_binding_sites_latest_Aug2022
Genome Assembly: hg38
Creation Date: 2022-09-01
Views: 1113
|
1662019200 |
1113 |
 |
Description: test
Author: ez176
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2022-09-01
Views: 772
|
1662019200 |
772 |
 |
Description: A session that isolates the 15 isoforms of SORBS1, a gene involved in insulin stimulation, and includes a custom track that isolates two of these isoforms. This session also includes the transcript expression GTEx track, which shows the frequencies of SORBS1 isoform expression in different types of tissue; Hence, one is able to see how each isoform of SORBS1 is expressed at different frequencies in each tissue type. The two isoforms highlighted within the custom track are great examples of isoform expression, as they are expressed in certain tissue types at vastly different rates.
Author: education
Session Name: hg19_SORBSct
Genome Assembly: hg19
Creation Date: 2022-08-30
Views: 969
|
1661846400 |
969 |
 |
Description: tracks for analysis of ERRg regulatory variants.
Author: agacita
Session Name: ERRg_basic
Genome Assembly: hg38
Creation Date: 2023-06-11
Views: 634
|
1686470400 |
634 |
 |
Description: The TBXT gene in its entirety. This gene is associated with the development of a tail and the way this gene is transcribed in hominoids results in their lack of an external tail. OMIM has two entries showing that variants on this gene are associated with spinal anomalies. For more information regarding this story, check out the Genome Browser education page: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_TBXTomim
Genome Assembly: hg19
Creation Date: 2022-08-30
Views: 1089
|
1661846400 |
1089 |
 |
Description: The squirrel monkey genome and the TBXT gene in its entirety. This gene is associated with the development of a tail and the way this gene is transcribed in hominoids results in their lack of an external tail. In monkeys, the inclusion of an exon in the mature transcript allows for formation of a tail. For more information regarding this story, check out the Genome Browser education page: https://genome.ucsc.edu/training/education
Author: education
Session Name: saiBol1_TBXT
Genome Assembly: saiBol1
Creation Date: 2022-08-24
Views: 935
|
1661328000 |
935 |
 |
Description: The TBXT gene in its entirety. This gene is associated with the development of a tail and the way this gene is transcribed in hominoids results in their lack of an external tail. Two Alu elements are highlighted; The yellow highlight (AluY) is unique to hominoids while the turquoise highlight (AluSx1) is found in all primates. The "Multiz Alignment" track illustrates which primates have the AluY element and which do not. For more information regarding this story, check out the Genome Browser education page: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_TBXTconservation
Genome Assembly: hg19
Creation Date: 2022-08-24
Views: 948
|
1661328000 |
948 |
 |
Description: The TBXT gene in its entirety. This gene is associated with the development of a tail and the way this gene is transcribed in hominoids results in their lack of an external tail. Two Alu elements are highlighted; The yellow highlight (AluY) is unique to hominoids while the turquoise highlight (AluSx1) is found in all primates. For more information regarding this story, check out the Genome Browser education page: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_TBXTalus
Genome Assembly: hg19
Creation Date: 2022-08-24
Views: 1214
|
1661328000 |
1214 |
 |
Description: The TBXT gene in its entirety. This gene is associated with the development of a tail and the way this gene is transcribed in hominoids results in their lack of an external tail. For more information regarding this story, visit the Genome Browser education page: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_TBXT
Genome Assembly: hg19
Creation Date: 2022-08-24
Views: 1043
|
1661328000 |
1043 |
 |
Description: BRCA1 gene region, showing CpG islands and DNA methylation. These regions influence gene expression.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_methylationAndIslands
Genome Assembly: hg19
Creation Date: 2022-08-19
Views: 1006
|
1660896000 |
1006 |
 |
Description: DNA methylation patterns with and without bi-sulfite treatment in BRCA1 region. These DNA modifications affect gene regulation.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_methylation
Genome Assembly: hg19
Creation Date: 2022-08-19
Views: 927
|
1660896000 |
927 |
 |
Description: Transcription factor binding sites and CpG island in BRCA1 region of human hg19 genome assembly.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_tfbs
Genome Assembly: hg19
Creation Date: 2022-08-19
Views: 1052
|
1660896000 |
1052 |
 |
Description: DMRs and DMPs in relation to H3K4me3 signals of Acute myeloid leukemia samples
Author: bacdao
Session Name: LAML
Genome Assembly: hg38
Creation Date: 2022-08-24
Views: 677
|
1661328000 |
677 |
 |
Description: POLN and HAUS3 share the first exon.
Author: Dongbo Ding
Session Name: POLN
Genome Assembly: hg19
Creation Date: 2022-08-06
Views: 813
|
1659772800 |
813 |
 |
Description: This session shows a portion of the ALDH2 gene, one of the two genes associated with alcohol intolerance in East Asian populations. The "GWAS" track is enabled and shows common nucleotide variants in the region and provides publications that further explore the consequences of a nucleotide change. Highlighted is rs671, which is the variant that causes “alcohol flush syndrome”; This condition is seen in approximately 30% to 50% of East Asian individuals.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_ALDH2gwas
Genome Assembly: hg19
Creation Date: 2022-07-21
Views: 992
|
1658390400 |
992 |
 |
Description: .....
Author: gperez2
Session Name: rn6_atp4
Genome Assembly: rn6
Creation Date: 2022-07-26
Views: 798
|
1658822400 |
798 |
 |
Description: Elys DamID in 16-18h Drosophila embryos.
Author: Shevelyov
Session Name: Elys_embryo DamID
Genome Assembly: dm3
Creation Date: 2022-12-14
Views: 737
|
1671004800 |
737 |
 |
Description: This session shows a portion of the ALDH2 gene, one of the two genes associated with alcohol intolerance in East Asian populations, in its entirety. The "Human Genome Diversity Project" (or "HGDP") track is enabled and marks variants that have different allele frequencies in individual populations. Clicking into a rsID provides specific population allele frequencies and a helpful visual aid. Highlighted is rs671, which is the variant that causes “alcohol flush syndrome”; This condition is seen in approximately 30% to 50% of East Asian individuals.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_ALDH2hgdp
Genome Assembly: hg19
Creation Date: 2022-07-21
Views: 868
|
1658390400 |
868 |
 |
Description: This session shows a portion of the ALDH2 gene, one of the two genes associated with alcohol intolerance in East Asian populations, in its entirety. The "Genome Aggregation Database" (or "gnomAD") track is enabled and marks variants that have different allele frequencies in individual populations. Clicking into a rsID provides specific population allele frequencies. Highlighted is rs671, which is the variant that causes “alcohol flush syndrome”; This condition is seen in approximately 30% to 50% of East Asian individuals. A click into the gnomAD track gives access to their population and other data.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_ALDH2gnomAD
Genome Assembly: hg19
Creation Date: 2022-07-21
Views: 996
|
1658390400 |
996 |
 |
Description: View of ARID1a gene with "CRISPR Targets" Data track enabled in pack mode. Find the story in UCSC's education module: https://genome.ucsc.edu/training/education.
Author: education
Session Name: crispr_fig3
Genome Assembly: hg38
Creation Date: 2022-08-28
Views: 955
|
1661673600 |
955 |
 |
Description: FGFR2 on the “UCSC Genes” and “NCBI Genes” tracks. FGFR2 is read on the opposite, negative strand, hence the leftward facing intron arrows. Different splicing events produce different isoforms of the gene.
Read more about splicing in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: fgfr2
Genome Assembly: hg19
Creation Date: 2022-02-24
Views: 1111
|
1645689600 |
1111 |
 |
Description: SNAPC1 gene shown with the first exon highlighted in orange.
Find a story about three reading frames in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: 3frame_fig1
Genome Assembly: hg38
Creation Date: 2022-06-27
Views: 1466
|
1656316800 |
1466 |
 |
Description: Peak file 'MCF7_E2-ethl.hpeak.out' comparing E2 and ethl-treated (control) samples.
Author: NSP
Session Name: MCF7_E2-ethl_Peaks_hg18
Genome Assembly: hg18
Creation Date: 2022-06-28
Views: 890
|
1656403200 |
890 |
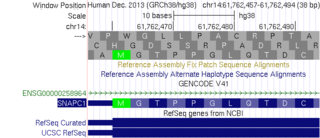 |
Description: A close-up of the first exon in the SNAPC1 gene. In this session, we can see the three different frames and the different amino acid sequences each frame contains. Notice how the frame with the methionine is the only frame read for the gene, as it encodes for the start of the first exon of the gene.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: 3frame_fig2
Genome Assembly: hg38
Creation Date: 2022-06-27
Views: 1188
|
1656316800 |
1188 |
 |
Description: Regions of genome detected in biofluids with at least 1 unique reads from samples present in the exRNA Atlas (exrna-atlas.org).
Author: elaplant
Session Name: hg19_exRNA_coverage_1
Genome Assembly: hg19
Creation Date: 2022-06-17
Views: 945
|
1655452800 |
945 |
 |
Description: Regions of genome detected in biofluids with at least 5 unique reads from samples present in the exRNA Atlas (exrna-atlas.org).
Author: elaplant
Session Name: hg19_exRNA_coverage_5
Genome Assembly: hg19
Creation Date: 2022-06-17
Views: 1207
|
1655452800 |
1207 |
 |
Description: This session shows the proximity of the LCT gene and two variants within the intron of the nearby MCM6 gene that affect the activity of the LCT gene -- causing persistence of lactase activity into adulthood. People with these variants can digest milk as adults. Without at least one of them, lactose intolerance results.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_LCT_MCM6
Genome Assembly: hg19
Creation Date: 2022-06-03
Views: 969
|
1654243200 |
969 |
 |
Description: This session highlights a single nucleotide change (G to A) with pathogenic consequences found within FOXP2, a gene associated with human speech and language comprehension. FOXP2's association with speech was discovered through this single nucleotide change first witnessed in the "KE family"; A pedigree in which almost half of the members for the past three generations have developed a speech disorder called verbal dyspraxia. The OMIM track is enabled and has a marker representing the nucleotide change found in the KE family pedigree. This G to A allele change results in a codon that corresponds to a different amino acid, histidine instead of the correct amino arginine (p.R553H), and results in a faulty protein which, as seen in the KE family, causes verbal dyspraxia.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_FOXP2p.R553H
Genome Assembly: hg19
Creation Date: 2022-05-15
Views: 936
|
1652601600 |
936 |
 |
Description: METTL3 gene zoomed in on the 3rd exon. The amino acid numbering is from right to left, denoting this exon/gene is on the bottom strand. Note the arrow at the upper left indicating that the DNA sequence at the top of the display window runs right to left. (If you are unable to see the amino acid numbering when viewing a gene at this scale, click on the gray sidebar box next to the gene, click on “Configure”, check the box labeled, “Show codon numbering”, and click “Apply”.)
Author: education
Session Name: hg38_revStrandZoom2
Genome Assembly: hg38
Creation Date: 2022-06-01
Views: 968
|
1654070400 |
968 |
 |
Description: METTL3 gene view zoomed in on the 3rd exon. The amino acid numbering is from right to left, denoting this exon/gene is on the bottom strand. Note the arrow at the upper left indicating that the DNA sequence at the top of the display window runs left to right -- which is the reverse complement of the sequence encoding the amino acids. (If you are unable to see the amino acid numbering when viewing a gene at this scale, click on the gray sidebar box next to the gene, click on “Configure”, check the box labeled, “Show codon numbering”, and click “Apply”.)
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg38_revStrandZoom
Genome Assembly: hg38
Creation Date: 2022-05-31
Views: 1001
|
1653984000 |
1001 |
 |
Description: MYC gene with exons and introns. The direction of the small intron arrows can be seen to go from left to right in the MYC gene, denoting the direction of the gene and signifying that the 5’ end is to the left and 3’ to the right (“top” strand).
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg38_MYC
Genome Assembly: hg38
Creation Date: 2022-05-31
Views: 1200
|
1653984000 |
1200 |
 |
Description: There are two amino acid differences in the FOXP2 gene between the genomes of humans and other primates that have been of interest lately. These amino acid changes are thought to be connected to why humans are capable of speech while chimpanzees and other apes are not. This session shows one of these amino acid differences using the Multiz Alignment track; Humans possess a serine (S) in the highlighted position while the other primates present on the track have an asparagine (N).
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_FOXP2fig4
Genome Assembly: hg19
Creation Date: 2022-05-29
Views: 902
|
1653811200 |
902 |
 |
Description: Editing for public
Author: QAtester
Session Name: hg38_highlight_v431_edit
Genome Assembly: hg38
Creation Date: 2022-05-26
Views: 916
|
1653552000 |
916 |
 |
Description: session
Author: QAtester3
Session Name: hg19_v431
Genome Assembly: hg19
Creation Date: 2022-05-24
Views: 907
|
1653379200 |
907 |
 |
Description: The PLP1 gene is shown with GTEx track, indicating the tissue specificity of gene expression. Note the high signal is brain tissues (yellow bars). The OMIM Allelic Variants track is expanded to full display mode, showing the phenotypes of variants in different locations in the gene.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_pelizaeus
Genome Assembly: hg19
Creation Date: 2022-05-15
Views: 1035
|
1652601600 |
1035 |
 |
Description: PLP1 gene and OMIM Allelic Variants track. Different variants result in different disease states (mouseover the OMIM Variants).
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_PLP1
Genome Assembly: hg19
Creation Date: 2022-05-15
Views: 1003
|
1652601600 |
1003 |
 |
Description: Highly conserved region in GRK4 gene. High PhyloP score for first and second nucleotides of codons reflects amino acid conservation through evolutionary time. Third base varies (wobble) because these amino acids have multiple codons -- the third base can be anything is free to diverge through time. PFAM track shows that this is a functionally significant region (protein kinase).
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_wobble2
Genome Assembly: hg19
Creation Date: 2022-05-12
Views: 973
|
1652342400 |
973 |
 |
Description: ChIP-seq, ATAC-seq, and RNA-seq count data from sorted porcine myeloid cells in duplicate.
Author: rcorbett
Session Name: susScr11.1_Myeloid_ChIP_ATAC
Genome Assembly: susScr11
Creation Date: 2022-05-06
Views: 943
|
1651824000 |
943 |
 |
Description: Quiz 1.
Author: jschifflin
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2023-09-05
Views: 568
|
1693900800 |
568 |
 |
Description: view of gene Rpl14 showing alternative splicing
Author: richardmott
Session Name: rn4.Rpl14
Genome Assembly: rn4
Creation Date: 2023-07-25
Views: 651
|
1690272000 |
651 |
 |
Description: view of gene Slc39a12 showing alternative splicing
Author: richardmott
Session Name: rn4.Slc39a12
Genome Assembly: rn4
Creation Date: 2023-07-25
Views: 586
|
1690272000 |
586 |
 |
Description:
Author: nannanzhao
Session Name: terc-ko
Genome Assembly: mm10
Creation Date: 2022-04-11
Views: 874
|
1649664000 |
874 |
 |
Description:
Author: davem
Session Name: midterm
Genome Assembly: hg38
Creation Date: 2022-04-06
Views: 890
|
1649232000 |
890 |
 |
Description: A session that isolates the 15 isoforms of SORBS1, a gene involved in insulin stimulation. This session also includes the two GTEx tracks, which illustrate frequencies of gene expression in different types of tissue.
Author: education
Session Name: hg19_SORBS1gtex
Genome Assembly: hg19
Creation Date: 2022-04-03
Views: 976
|
1648972800 |
976 |
 |
Description: A session that isolates the 15 isoforms of SORBS1, a gene involved in insulin stimulation. This session is utilized on the Education page in the "splice variants" document.
Author: education
Session Name: hg19_SORBS1
Genome Assembly: hg19
Creation Date: 2022-04-03
Views: 1081
|
1648972800 |
1081 |
 |
Description: There are two amino acid differences in the FOXP2 gene between the genomes of humans and other primates that have been of interest lately. These amino acid changes are thought to be connected to why humans are capable of speech while chimpanzees and other apes are not. This session shows one of these amino acid differences using the Multiz Alignment track; Humans possess an asparagine (N) in the highlighted position while the other primates present on the track have a threonine (T).
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_FOXP2fig3
Genome Assembly: hg19
Creation Date: 2022-05-15
Views: 915
|
1652601600 |
915 |
 |
Description: RHO hg19
Author: Chaffikm
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2023-01-26
Views: 688
|
1674720000 |
688 |
 |
Description:
Author: ZC
Session Name: ATAC
Genome Assembly: hg38
Creation Date: 2022-03-25
Views: 976
|
1648195200 |
976 |
 |
Description:
Author: EsterCastillo
Session Name: Paneles_VHIO_bed
Genome Assembly: hg19
Creation Date: 2022-03-25
Views: 904
|
1648195200 |
904 |
 |
Description: Tracks from GEO GSE22104
Samples:
GSM550303 Stat4WTTh1
GSM550304 Stat4KOTh1
GSM550305 H3K4me3WTTh1
GSM550306 H3K4me3 S4KOTh1
GSM550307 H3K27me3WTTh1
GSM550308 H3K27me3 S4KOTh1
content.txt files and bedgraph files
Author: twyss
Session Name: Chip_Stat4_Th1_Wei
Genome Assembly: mm9
Creation Date: 2024-12-13
Views: 260
|
1734076800 |
260 |
 |
Description: This session shows a short match on the UTR sequence for a gene, acugcugagcugggag. If you click below the black box, into the gene page for TGDS, you can then click an "RNA Structure" link. This will bring you to a section where you can then see for the 5' UTR a "Picture" link and see how this RNA fold structure may arrange.
Author: QAtester
Session Name: RNA_plot
Genome Assembly: hg38
Creation Date: 2022-05-03
Views: 1084
|
1651564800 |
1084 |
 |
Description: CHip
Author: xiaopei
Session Name: Chip_seq_B
Genome Assembly: hg38
Creation Date: 2024-07-01
Views: 332
|
1719820800 |
332 |
 |
Description: Review in this mailing list question, https://groups.google.com/a/soe.ucsc.edu/g/genome/c/y3evkDazY2o/m/0WjoQD63BQAJ, the "How to start from scratch to find tracks" and "How to view data in the region of interest and adjust display" sections and instead of using hg38 go to "GRCm38/mm10" and then use "TF" in the track search step. Set the "JASPAR Transcription Factors" track on mm10 to "pack" and then search "Mef2C" and select "Mef2c (ENSMUST00000198199.4) at chr13:83504034-83663343" from the search results, and then zoom out "1.5x" as well. Next click into the grey box for the "JASPAR Transcription Factors Track Settings" and click the "Schema" link on the far right. By clicking the Schema link you will see the fields for the "Primary Table: jaspar2022" and from this page you can use the Tools menu to arrive at the Table Browser, so this table will be automatically selected for you. Here click the "create" button next to "filter:" and for the "score is" field set it from "ignored" to ">" for greater than and put in a value such as 700 and then click submit. Then put in the "output filename:" field a name like "mm10Jaspar_Mef2c_score700.csv" and be sure that you click the button next to "csv (for excel)" and then click "get output" to download the results. Open the resulting file in Excel.
Author: brianlee
Session Name: Shiaoching
Genome Assembly: mm10
Creation Date: 2022-03-16
Views: 884
|
1647417600 |
884 |
 |
Description: Mm10 browser session associated with our manuscript "Integration of multimodal data in the developing tooth reveals candidate regulatory loci driving human odontogenic phenotypes", Frontiers in Dental Medicine, 2022
Author: emmawentworth
Session Name: mm10_all_18state
Genome Assembly: mm10
Creation Date: 2022-03-08
Views: 1723
|
1646726400 |
1723 |
 |
Description:
Author: hy123
Session Name: hg38 heart 1
Genome Assembly: hg38
Creation Date: 2022-03-07
Views: 896
|
1646640000 |
896 |
 |
Description:
Author: asaenz.13
Session Name: Genomics group 7 - final project
Genome Assembly: hg19
Creation Date: 2022-03-04
Views: 923
|
1646380800 |
923 |
 |
Description:
Author: YKehayova
Session Name: hg19 ATAC-seq 01.03.22
Genome Assembly: hg19
Creation Date: 2022-03-01
Views: 935
|
1646121600 |
935 |
 |
Description:
Author: aytyuh
Session Name: new
Genome Assembly: mm10
Creation Date: 2022-02-16
Views: 955
|
1644998400 |
955 |
 |
Description:
Author: JFormosa
Session Name: 2-15-22 Hwk6pt1
Genome Assembly: hg19
Creation Date: 2022-02-15
Views: 900
|
1644912000 |
900 |
 |
Description: This session shows a portion of the ALDH2 gene, one of the two genes associated with alcohol intolerance in East Asian populations. The "OMIM" track is enabled and shows common nucleotide variants in the region and provides information regarding the consequences of a nucleotide change. Highlighted is rs671, which is the variant that causes “alcohol flush syndrome”; This condition is seen in approximately 30% to 50% of East Asian individuals. Clicking into the OMIM track provides access to details hosted there.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_ALDH2omim
Genome Assembly: hg19
Creation Date: 2022-07-21
Views: 920
|
1658390400 |
920 |
 |
Description: This session shows the ALDH2 gene, one of the two genes associated with alcohol intolerance in East Asian populations, in its entirety. The "dbSNP 150" track is enabled and shows common nucleotide variants in the region. Highlighted is rs671, which is the variant that causes “alcohol flush syndrome”; This condition is seen in approximately 30% to 50% of East Asian individuals. A smaller rsID number corresponds to an earlier entry, meaning that rs671 is one of the earliest uploaded into NCBIs dbSnp database.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_ALDH2dbSNP150
Genome Assembly: hg19
Creation Date: 2022-07-21
Views: 980
|
1658390400 |
980 |
 |
Description:
Author: Chilliard
Session Name: hg19 - assignment 5
Genome Assembly: hg19
Creation Date: 2022-02-15
Views: 1217
|
1644912000 |
1217 |
 |
Description:
Author: JFormosa
Session Name: 2-14-22 Hwk 5
Genome Assembly: hg19
Creation Date: 2022-02-14
Views: 920
|
1644825600 |
920 |
 |
Description: Desiccation-induced inflammation and ocular surface disease is suppressed by terminal state MRC1 conjunctival macrophages.
Author: zhenzuo2
Session Name: MRC1_conjunctival_macrophages
Genome Assembly: mm10
Creation Date: 2024-04-23
Views: 453
|
1713859200 |
453 |
 |
Description:
Author: himtiffant
Session Name: p1 t1
Genome Assembly: mm10
Creation Date: 2022-02-09
Views: 1145
|
1644393600 |
1145 |
 |
Description:
Author: helenajara10
Session Name: mm10w/ Dock6
Genome Assembly: mm10
Creation Date: 2022-02-09
Views: 925
|
1644393600 |
925 |
 |
Description:
Author: aytyuh
Session Name: task 1
Genome Assembly: mm10
Creation Date: 2022-02-09
Views: 895
|
1644393600 |
895 |
 |
Description:
Author: helenajara10
Session Name: mm10w/interaction
Genome Assembly: mm10
Creation Date: 2022-02-09
Views: 881
|
1644393600 |
881 |
 |
Description:
Author: Jcotney
Session Name: TFAP2_Enhancer_KO_Session
Genome Assembly: hg38
Creation Date: 2022-02-08
Views: 1012
|
1644307200 |
1012 |
 |
Description:
Author: chunmei1991
Session Name: ChIPseq_peak_profile_Weilab
Genome Assembly: hg19
Creation Date: 2022-02-06
Views: 1059
|
1644134400 |
1059 |
 |
Description: Close view of the ARID1a gene with the "CRISPR Targets" data track enabled. Filter is set to 90 to show only the best sites. Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: crispr_fig5
Genome Assembly: hg38
Creation Date: 2022-08-28
Views: 908
|
1661673600 |
908 |
 |
Description: This session isolates a nonsense single nucleotide variant (rs886040510), highlighted in turquoise, found on BRCA2 which is a gene associated with breast cancer. The "dbSNP" track has information regarding this variant. This variant is an A to T change and is known to have pathogenic consequences. The "ClinVar" track confirms rs886040510's pathogenicity with a red box and bead. Clicking onto either of these markings will navigate you to a details page that provides more information.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_BRCA2stopclinvar
Genome Assembly: hg19
Creation Date: 2022-02-03
Views: 907
|
1643875200 |
907 |
 |
Description: This session isolates a nonsense single nucleotide variant (rs886040510), highlighted in turquoise, found on BRCA2 which is a gene associated with breast cancer. The dbSNP track has information regarding this variant. This variant is an A to T change and is known to have pathogenic consequences.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_BRCA2stopgain
Genome Assembly: hg19
Creation Date: 2022-02-03
Views: 1152
|
1643875200 |
1152 |
 |
Description: This session shows a synonymous variant found on BRCA2, a gene associated with breast cancer. The nucleotide change does result in a changed amino acid. The variant in question is highlighted in turquoise. Two dbSNP tracks are enabled and provide more information about the variant.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_BRCA2synon
Genome Assembly: hg19
Creation Date: 2022-02-03
Views: 946
|
1643875200 |
946 |
 |
Description: This session shows a frameshift variant (highlighted in turquoise) on BRCA2, a gene associated with breast cancer. The variant is a deletion of two nucleotides, C and T. This deletion results in a change in the reading frame. Two dbSNP tracks are enabled and provide more information on the variant.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_BRCA2frameshift
Genome Assembly: hg19
Creation Date: 2022-02-03
Views: 930
|
1643875200 |
930 |
 |
Description: This session shows a frameshift variant (highlighted in turquoise, but looks pale green under the yellow highlight) on BRCA2, a gene associated with breast cancer. The variant is a deletion of two nucleotides, C and T. This deletion results in a change in the reading frame. The pale yellow highlight shows this change, AGT being the new codon after the C and T nucleotides (pale green) are deleted. This changes the reading frame from the second frame to the first frame shown on the Base Position track, which is set to full in order to visualize three potential reading frames. The red highlight marks the stop codon of this new reading frame. Therefore, this frameshift variant results in a premature stop codon. The dbSNP track is enabled and provides more information on the variant.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_BRCA2frames
Genome Assembly: hg19
Creation Date: 2022-02-03
Views: 907
|
1643875200 |
907 |
 |
Description: This session shows a missense variant, highlighted in turquoise, found on BRCA2. This single nucleotide variant (SNV) changes the resulting amino acid; The original codon "CAT" codes for histidine while an individual with the variant in question would possess either the codon "CAA" or "CAG", coding for glutamine. This means the resulting polypeptide chain would be impacted. However, ClinVar's data does not label this variant as pathogenic.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_BRCA2missense
Genome Assembly: hg19
Creation Date: 2022-02-03
Views: 936
|
1643875200 |
936 |
 |
Description: This session shows a portion of BRCA2 (exon 11), a gene associated with breast cancer. Two dbSNP tracks are enabled and mark known variants in the region displayed. Red boxes represent variants that disrupt effective protein production (missense), while green boxes indicate that the variant does not change the resulting amino acid (synonymous). Some of these missense variants have pathogenic consequences, causing breast cancer.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_BRCA2variants
Genome Assembly: hg19
Creation Date: 2022-02-03
Views: 1130
|
1643875200 |
1130 |
 |
Description: The genes HBB and HBD are both expressed in red blood cells, but HBB is also expressed in many other tissues, whereas HBD is expressed in red cells almost exclusively.
Author: helenajara10
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2022-02-03
Views: 1360
|
1643875200 |
1360 |
 |
Description:
Author: Kaushik
Session Name: PHOX2B
Genome Assembly: hg18
Creation Date: 2022-02-05
Views: 966
|
1644048000 |
966 |
 |
Description:
Author: pwjeffries
Session Name: hg38Covert
Genome Assembly: hg38
Creation Date: 2022-02-02
Views: 935
|
1643788800 |
935 |
 |
Description:
Author: katerynamarku
Session Name: Fall19 Yeast test (KM)
Genome Assembly: sacCer3
Creation Date: 2022-01-31
Views: 970
|
1643616000 |
970 |
 |
Description:
Author: pham4tt
Session Name: Spring20 Yeat Test TM,KM,EL
Genome Assembly: sacCer3
Creation Date: 2022-01-31
Views: 880
|
1643616000 |
880 |
 |
Description:
Author: tomgo
Session Name: mm10- enhancers
Genome Assembly: mm10
Creation Date: 2022-01-31
Views: 903
|
1643616000 |
903 |
 |
Description:
Author: jos200a
Session Name: Fall 2022 Yeast Test JEAA
Genome Assembly: sacCer3
Creation Date: 2022-01-30
Views: 897
|
1643529600 |
897 |
 |
Description:
Author: Cliffford081915
Session Name: Spring2022 Yeast Test(JDC)
Genome Assembly: sacCer3
Creation Date: 2022-01-30
Views: 892
|
1643529600 |
892 |
 |
Description:
Author: garberla
Session Name: CGEMS yeast LG test
Genome Assembly: sacCer3
Creation Date: 2022-01-26
Views: 1019
|
1643184000 |
1019 |
 |
Description:
Author: ajaycox
Session Name: CGEMS yeast (AJ) test
Genome Assembly: sacCer3
Creation Date: 2022-01-26
Views: 935
|
1643184000 |
935 |
 |
Description:
Author: espinokb
Session Name: CGEMS yeast KE test
Genome Assembly: sacCer3
Creation Date: 2022-01-26
Views: 936
|
1643184000 |
936 |
 |
Description:
Author: CherryLab
Session Name: Nuclear_EyeBrowser_TrackHub
Genome Assembly: hg38
Creation Date: 2021-11-01
Views: 1572
|
1635753600 |
1572 |
 |
Description:
Author: aytyuh
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2022-02-09
Views: 1144
|
1644393600 |
1144 |
 |
Description:
Author: YKehayova
Session Name: hg19 SBNO1 project
Genome Assembly: hg19
Creation Date: 2022-01-20
Views: 964
|
1642665600 |
964 |
 |
Description:
Author: Pey.davoodi
Session Name: Optional
Genome Assembly: hg19
Creation Date: 2022-02-07
Views: 885
|
1644220800 |
885 |
 |
Description:
Author: Pey.davoodi
Session Name: 4-genes-workshop
Genome Assembly: hg19
Creation Date: 2022-02-07
Views: 921
|
1644220800 |
921 |
 |
Description:
Author: YKehayova
Session Name: hg19 ATAC peaks COLGALT2 promoter
Genome Assembly: hg19
Creation Date: 2022-01-19
Views: 1022
|
1642579200 |
1022 |
 |
Description: ChIP, ATAC, and RNA-seq counts for sorted porcine myeloid cells in duplicate, along with H3K27ac and H3K4me3 data from other cell types for comparison.
Author: rcorbett
Session Name: susScr11_Myeloid_ChIP_ATAC_comparison
Genome Assembly: susScr11
Creation Date: 2022-05-06
Views: 964
|
1651824000 |
964 |
 |
Description: CAG repeats in the HTT gene. Expansion of the CAG repeats leads to Huntington's disease.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: htt_main
Genome Assembly: hg19
Creation Date: 2022-01-13
Views: 1185
|
1642060800 |
1185 |
 |
Description: A session highlighting two single-nucleotide variants found in introns 9 and 13 of the MCM6 gene, located upstream from the LCT gene, that impact whether or not an individual is lactase persistent as an adult. Both the OMIM and GWAS tracks have marked these variants as phenotypically relevant to the appearance of lactase persistence.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_LCT
Genome Assembly: hg19
Creation Date: 2021-12-25
Views: 1418
|
1640419200 |
1418 |
 |
Description:
Author: dxliucmu
Session Name: hg38_ ChIP with H3K4me1 in HC vs SCZ (one pair))
Genome Assembly: hg38
Creation Date: 2022-01-09
Views: 892
|
1641715200 |
892 |
 |
Description:
Author: JFormosa
Session Name: Hwk6Pt1
Genome Assembly: hg19
Creation Date: 2022-02-22
Views: 884
|
1645516800 |
884 |
 |
Description: UCLA 2022 workshop -- custom tracks Alus from dbRIP not in rmsk; CAT gene with two names, some SNPs from 151 missense and intron
Author: Example
Session Name: hg19_snp_CAT_Alus
Genome Assembly: hg19
Creation Date: 2022-01-20
Views: 944
|
1642665600 |
944 |
 |
Description:
Author: ani_davis
Session Name: Spring22 Yeast Test AND
Genome Assembly: sacCer3
Creation Date: 2022-01-31
Views: 914
|
1643616000 |
914 |
 |
Description: IPMK intron Author: Mustafi P
Session Name: ATAC-seqPC4
Genome Assembly: hg19
Author: pallabidgp123
Session Name: ATAC-seq6
Genome Assembly: hg19
Creation Date: 2021-12-25
Views: 1003
|
1640419200 |
1003 |
 |
Description: ATG5 intron Author: Mustafi P
Session Name: ATAC-seqPC4
Genome Assembly: hg19
Author: pallabidgp123
Session Name: ATAC-seq4
Genome Assembly: hg19
Creation Date: 2021-12-23
Views: 1024
|
1640246400 |
1024 |
 |
Description: MAP3K7 intron Author: Mustafi P
Session Name: ATAC-seq2
Genome Assembly: hg19
Author: pallabidgp123
Session Name: ATAC-seq2
Genome Assembly: hg19
Creation Date: 2021-12-23
Views: 1067
|
1640246400 |
1067 |
 |
Description: UVRAG promoter-TSS Author: Mustafi P
Session Name: ATAC-seqPC4
Genome Assembly: hg19
Author: pallabidgp123
Session Name: ATAC-seq1
Genome Assembly: hg19
Creation Date: 2021-12-23
Views: 1089
|
1640246400 |
1089 |
 |
Description: IPMK intron Author: Mustafi P
Session Name: ATAC-seqPC4
Genome Assembly: hg19
Author: pallabidgp123
Session Name: ATAC-seq PC4
Genome Assembly: hg19
Creation Date: 2021-12-23
Views: 952
|
1640246400 |
952 |
 |
Description: IPMK intron Author: Mustafi P
Session Name: ATAC-seq5
Genome Assembly: hg19
Author: pallabidgp123
Session Name: ATAC-seq5
Genome Assembly: hg19
Creation Date: 2021-12-23
Views: 1010
|
1640246400 |
1010 |
 |
Description: YEATS2 intron TSS Author: Mustafi P
Session Name: ATAC-seq3
Genome Assembly: hg19
Author: pallabidgp123
Session Name: ATAC-seq3
Genome Assembly: hg19
Creation Date: 2021-12-23
Views: 983
|
1640246400 |
983 |
 |
Description: Close-up view if transition zone between a coding and a non-coding region of the virus. Note the High conservation in the PhyloP track in the protein-coding region.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: ebolav_pconserv
Genome Assembly: eboVir3
Creation Date: 2021-12-22
Views: 844
|
1640160000 |
844 |
 |
Description: Ebola virus VP24 gene with PhyloP track showing high conservation in the protein-coding region among 160+ isolates.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: ebolav_phylop
Genome Assembly: eboVir3
Creation Date: 2021-12-22
Views: 1053
|
1640160000 |
1053 |
 |
Description: Ebola virus genome showing vp24 gene region. The PhyloP track shows that the 100+ strains of ebola and Marburg have conservation in the protein-coding region of the gene. Pink color indicates a positive value above the display level in the track.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: ebolav_vp24
Genome Assembly: eboVir3
Creation Date: 2021-12-22
Views: 891
|
1640160000 |
891 |
 |
Description: Zoomed-in view of ebolavirus L protein showing serine codon in the reference that is different in many islolates distantly related to it (lower part of figure).
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: ebolav_codon
Genome Assembly: eboVir3
Creation Date: 2021-12-22
Views: 883
|
1640160000 |
883 |
 |
Description: Full genome of the ebola virus showing a blat alignment of the Marburg virus.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: ebolav_blatsearch
Genome Assembly: eboVir3
Creation Date: 2021-12-22
Views: 901
|
1640160000 |
901 |
 |
Description: Full genome of ebola virus showing proteins in the top track and conservation among several outbreaks in the lower tracks.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: ebolav_mainsession
Genome Assembly: eboVir3
Creation Date: 2021-12-22
Views: 928
|
1640160000 |
928 |
 |
Description: Visualization of the BRCA1 Gene, CpG islands and DNA methylations in different cell lines using the bisulfite Seq and Methyl 450k method.
Author: art_03m
Session Name: hg19: Methylation & CpG islands in BRCA1 gene
Genome Assembly: hg19
Creation Date: 2021-12-18
Views: 637
|
1639814400 |
637 |
 |
Description:
Author: adelaidetovar
Session Name: apoa2
Genome Assembly: hg38
Creation Date: 2021-12-13
Views: 903
|
1639382400 |
903 |
 |
Description:
Author: SKellaway
Session Name: dnFOSforSNPs
Genome Assembly: hg38
Creation Date: 2021-12-08
Views: 946
|
1638950400 |
946 |
 |
Description:
Author: Schne2lk
Session Name: hg38 NRL multiomics
Genome Assembly: hg38
Creation Date: 2021-12-07
Views: 927
|
1638864000 |
927 |
 |
Description:
Author: marielgr
Session Name: ACSS2_ChIPseq
Genome Assembly: mm39
Creation Date: 2021-12-17
Views: 897
|
1639728000 |
897 |
 |
Description: Aging MapR data in photoreceptor neurons. Loaded bigwig track correspond to the averaged signal for each aging time point generated from three independent biological replicates
Author: juanjaureguilozano
Session Name: dm6_agingMapR
Genome Assembly: dm6
Creation Date: 2021-12-01
Views: 9634
|
1638345600 |
9634 |
 |
Description: A pipeline for predicting cardiac gene network components using cis-regulatory elements (CREs)
Manuscript: Hieu T. Nim*, Louis Dang*, Harshini Thiyagarajah*, Daniel Bakopoulos, Michael See, Natalie Charitakis, Tennille Sibbritt, Michael P. Eichenlaub, Stuart K. Archer, Nicolas Fossat, Richard E. Burke, Patrick P. L. Tam, Coral G. Warr‡, Travis K. Johnson‡#, Mirana Ramialison‡#. "Predicting genes essential for heart development irrespective of their spatial expression pattern". Genome Biology (in press). (*: co-first authors; ‡: co-senior authors; #: corresponding authors)
Author: nimt0001
Session Name: CardiacNetworkComponentPredictor
Genome Assembly: mm9
Creation Date: 2021-11-30
Views: 1211
|
1638259200 |
1211 |
 |
Description:
Author: latimeaa
Session Name: Hg38 HISAT2 Retina tracks
Genome Assembly: hg38
Creation Date: 2021-11-30
Views: 946
|
1638259200 |
946 |
 |
Description:
Author: Eldar
Session Name: SNV
Genome Assembly: hg38
Creation Date: 2021-02-16
Views: 1325
|
1613462400 |
1325 |
 |
Description:
Author: Eldar
Session Name: CNV
Genome Assembly: hg38
Creation Date: 2021-02-25
Views: 1645
|
1614240000 |
1645 |
 |
Description:
Author: aigul
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2021-11-18
Views: 1013
|
1637222400 |
1013 |
 |
Description:
Author: Wen-Cheng
Session Name: Liver_APOA5p
Genome Assembly: hg19
Creation Date: 2020-11-04
Views: 1617
|
1604476800 |
1617 |
 |
Description:
Author: zunpengliu66
Session Name: WT H3K9me3 ChIP-seq
Genome Assembly: hg38
Creation Date: 2021-11-11
Views: 895
|
1636617600 |
895 |
 |
Description:
Author: randa2jl
Session Name: Human retina multiomics
Genome Assembly: hg38
Creation Date: 2021-11-16
Views: 915
|
1637049600 |
915 |
 |
Description:
Author: young_researcher
Session Name: hg19_intersection
Genome Assembly: hg19
Creation Date: 2021-11-07
Views: 1240
|
1636272000 |
1240 |
 |
Description:
Author: young_researcher
Session Name: hg19_H3F3A
Genome Assembly: hg19
Creation Date: 2021-11-07
Views: 1077
|
1636272000 |
1077 |
 |
Description:
Author: vladislav
Session Name: intersect_with_DeepZ
Genome Assembly: hg19
Creation Date: 2021-11-06
Views: 935
|
1636185600 |
935 |
 |
Description:
Author: vladislav
Session Name: H3F3A.merge
Genome Assembly: hg19
Creation Date: 2021-11-06
Views: 1003
|
1636185600 |
1003 |
 |
Description:
Author: vladislav
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2021-11-06
Views: 1082
|
1636185600 |
1082 |
 |
Description:
Author: megallegos
Session Name: Independent Project Link
Genome Assembly: hg19
Creation Date: 2021-11-05
Views: 1327
|
1636099200 |
1327 |
 |
Description:
Author: randa2jl
Session Name: PDE6A
Genome Assembly: mm9
Creation Date: 2021-11-04
Views: 972
|
1636012800 |
972 |
 |
Description:
Author: Schne2lk
Session Name: mm9 RCVRN
Genome Assembly: mm9
Creation Date: 2021-11-04
Views: 953
|
1636012800 |
953 |
 |
Description:
Author: Schne2lk
Session Name: mm9 HTT
Genome Assembly: mm9
Creation Date: 2021-11-04
Views: 1445
|
1636012800 |
1445 |
 |
Description:
Author: Schne2lk
Session Name: mm9 thrb
Genome Assembly: mm9
Creation Date: 2021-11-04
Views: 962
|
1636012800 |
962 |
 |
Description:
Author: randa2jl
Session Name: wt/nrl CBRs
Genome Assembly: mm9
Creation Date: 2021-11-04
Views: 1331
|
1636012800 |
1331 |
 |
Description:
Author: laarne
Session Name: CHIP SEQ ACTUAL SESSION CBR
Genome Assembly: mm9
Creation Date: 2021-11-04
Views: 959
|
1636012800 |
959 |
 |
Description:
Author: Schne2lk
Session Name: mm9 PDE6A
Genome Assembly: mm9
Creation Date: 2021-11-04
Views: 983
|
1636012800 |
983 |
 |
Description:
Author: adityabhagwate
Session Name: bw_upload
Genome Assembly: hg19
Creation Date: 2021-11-03
Views: 1013
|
1635926400 |
1013 |
 |
Description:
Author: ashleywilkins
Session Name: DSP
Genome Assembly: hg19
Creation Date: 2021-11-02
Views: 1471
|
1635840000 |
1471 |
 |
Description:
Author: sweatbr
Session Name: MYBPC1
Genome Assembly: hg19
Creation Date: 2021-11-02
Views: 1003
|
1635840000 |
1003 |
 |
Description:
Author: sweatbr
Session Name: DSP
Genome Assembly: hg19
Creation Date: 2021-11-02
Views: 1216
|
1635840000 |
1216 |
 |
Description:
Author: randa2jl
Session Name: MYBPC1
Genome Assembly: hg19
Creation Date: 2021-11-02
Views: 1475
|
1635840000 |
1475 |
 |
Description:
Author: ashleywilkins
Session Name: Cardiomyopathy
Genome Assembly: hg19
Creation Date: 2021-11-02
Views: 1071
|
1635840000 |
1071 |
 |
Description:
Author: Schne2lk
Session Name: hg19 DSP
Genome Assembly: hg19
Creation Date: 2021-11-02
Views: 923
|
1635840000 |
923 |
 |
Description:
Author: randa2jl
Session Name: hg19-RHO custom track
Genome Assembly: hg19
Creation Date: 2021-11-02
Views: 906
|
1635840000 |
906 |
 |
Description:
Author: Schne2lk
Session Name: hg19 MYBPC1
Genome Assembly: hg19
Creation Date: 2021-11-02
Views: 956
|
1635840000 |
956 |
 |
Description:
Author: sweatbr
Session Name: RIDLEY
Genome Assembly: hg38
Creation Date: 2021-11-04
Views: 994
|
1636012800 |
994 |
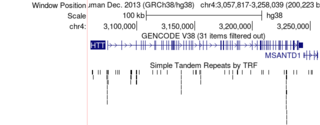 |
Description:
Author: randa2jl
Session Name: Fall21 HTT gene Huntingtons
Genome Assembly: hg38
Creation Date: 2021-10-19
Views: 1123
|
1634630400 |
1123 |
 |
Description:
Author: Schne2lk
Session Name: hg38 HTT
Genome Assembly: hg38
Creation Date: 2021-10-19
Views: 965
|
1634630400 |
965 |
 |
Description:
Author: whaleycn
Session Name: HTT
Genome Assembly: hg38
Creation Date: 2021-10-19
Views: 983
|
1634630400 |
983 |
 |
Description:
Author: JuliaLaz
Session Name: HTT
Genome Assembly: hg38
Creation Date: 2021-10-18
Views: 1034
|
1634544000 |
1034 |
 |
Description:
Author: ashleywilkins
Session Name: hg38 HTT GENE
Genome Assembly: hg38
Creation Date: 2021-10-17
Views: 1213
|
1634457600 |
1213 |
 |
Description:
Author: CherryLab
Session Name: VISIONS_TrackHub
Genome Assembly: hg38
Creation Date: 2021-10-15
Views: 1155
|
1634284800 |
1155 |
 |
Description: This session includes two custom tracks on chromosome 22, spanning the region 20100000-20140000. The first track, named 'spacer', displays blue ticks every 10,000 bases. The second track, named 'even', shows red ticks every 100 bases, skipping 100 bases between each tick. The 'even' track also includes labels for the first three ticks: 'first', 'second', and 'third'. These custom tracks demonstrate the use of BED format for visualizing specific genomic intervals and annotations.
Author: Parth Doshi
Session Name: Custom Track Assignment
Genome Assembly: hg38
Creation Date: 2024-11-06
Views: 220
|
1730880000 |
220 |
 |
Description:
Author: yoni.arndt
Session Name: ex.1 RAC1
Genome Assembly: hg38
Creation Date: 2021-10-15
Views: 1011
|
1634284800 |
1011 |
 |
Description:
Author: randa2jl
Session Name: Fall21 HGD gene
Genome Assembly: hg38
Creation Date: 2021-10-14
Views: 949
|
1634198400 |
949 |
 |
Description:
Author: sweatbr
Session Name: chr3chimp
Genome Assembly: hg38
Creation Date: 2021-10-12
Views: 1078
|
1634025600 |
1078 |
 |
Description:
Author: randa2jl
Session Name: Fall21 HumanChr2 JennaRandall
Genome Assembly: hg38
Creation Date: 2021-10-12
Views: 1210
|
1634025600 |
1210 |
 |
Description:
Author: JFormosa
Session Name: Hwk6Pt4b
Genome Assembly: hg19
Creation Date: 2022-02-22
Views: 872
|
1645516800 |
872 |
 |
Description:
Author: JFormosa
Session Name: Hwk6Pt4a
Genome Assembly: hg19
Creation Date: 2022-02-22
Views: 901
|
1645516800 |
901 |
 |
Description:
Author: mjliddi16
Session Name: hg19_assignment6
Genome Assembly: hg19
Creation Date: 2022-02-22
Views: 1271
|
1645516800 |
1271 |
 |
Description: BINF6310
Author: Rahulprasad Selvaraj
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2023-11-08
Views: 472
|
1699430400 |
472 |
 |
Description:
Author: tboisjol
Session Name: TB-98358724-MEDG580-UBC-2020
Genome Assembly: hg38
Creation Date: 2021-10-08
Views: 1199
|
1633680000 |
1199 |
 |
Description:
Author: millerh1@livemail.uthscsa.edu
Session Name: RLBase
Genome Assembly: hg38
Creation Date: 2021-10-06
Views: 2585
|
1633507200 |
2585 |
 |
Description:
Author: millerh1@livemail.uthscsa.edu
Session Name: RLBase_slow
Genome Assembly: hg38
Creation Date: 2021-10-05
Views: 1136
|
1633420800 |
1136 |
 |
Description:
Author: kkaczor
Session Name: tamer_hiv_atac
Genome Assembly: hg38
Creation Date: 2021-09-30
Views: 1252
|
1632988800 |
1252 |
 |
Description:
Author: kkaczor
Session Name: tamer_hiv_rna
Genome Assembly: hg38
Creation Date: 2021-09-29
Views: 1202
|
1632902400 |
1202 |
 |
Description:
Author: laarne
Session Name: hg38 HTT gene
Genome Assembly: hg38
Creation Date: 2021-10-19
Views: 1020
|
1634630400 |
1020 |
 |
Description:
Author: areynolds
Session Name: hg38 HBB-LCR
Genome Assembly: hg38
Creation Date: 2021-09-20
Views: 1152
|
1632124800 |
1152 |
 |
Description:
Author: jschlachetzki
Session Name: 20210917_4DBSM_Schlachetzki
Genome Assembly: hg19
Creation Date: 2021-09-17
Views: 979
|
1631865600 |
979 |
 |
Description:
Author: layla13
Session Name: TSA2R38 Group #4
Genome Assembly: hg19
Creation Date: 2021-09-16
Views: 914
|
1631779200 |
914 |
 |
Description:
Author: Thijs
Session Name: Wnt_human_NGS_20210914_V2
Genome Assembly: hg38
Creation Date: 2021-09-14
Views: 1194
|
1631606400 |
1194 |
 |
Description:
Author: mallorycunningham
Session Name: TASR38 group3
Genome Assembly: hg19
Creation Date: 2021-09-12
Views: 1150
|
1631433600 |
1150 |
 |
Description:
Author: noycohen
Session Name: HepG2 and K562 Methylation
Genome Assembly: hg38
Creation Date: 2021-09-12
Views: 1383
|
1631433600 |
1383 |
 |
Description:
Author: msadams
Session Name: A549
Genome Assembly: hg38
Creation Date: 2021-09-28
Views: 939
|
1632816000 |
939 |
 |
Description:
Author: xuxing
Session Name: multi-Paired-seq
Genome Assembly: hg38
Creation Date: 2021-10-31
Views: 976
|
1635667200 |
976 |
 |
Description:
Author: jessicagarth
Session Name: TAS2R38 group4
Genome Assembly: hg19
Creation Date: 2021-09-08
Views: 1137
|
1631088000 |
1137 |
 |
Description:
Author: hayes22
Session Name: Fall21 Yeast Test JEH
Genome Assembly: sacCer3
Creation Date: 2021-09-08
Views: 1135
|
1631088000 |
1135 |
 |
Description:
Author: SamanthaBusch
Session Name: Fall21 Yeast test SIB
Genome Assembly: sacCer3
Creation Date: 2021-09-07
Views: 1192
|
1631001600 |
1192 |
 |
Description:
Author: Siddeldj
Session Name: Fall19 yeast test DJS
Genome Assembly: sacCer3
Creation Date: 2021-09-07
Views: 1157
|
1631001600 |
1157 |
 |
Description:
Author: Albert Cheng
Session Name: hg38_ZFP18_1
Genome Assembly: hg38
Creation Date: 2021-09-07
Views: 1199
|
1631001600 |
1199 |
 |
Description:
Author: Albert Cheng
Session Name: hg38_Cas9_1_0_0_0
Genome Assembly: hg38
Creation Date: 2021-09-07
Views: 1108
|
1631001600 |
1108 |
 |
Description:
Author: Albert Cheng
Session Name: hg38_ZFP18_1_0
Genome Assembly: hg38
Creation Date: 2021-09-07
Views: 1185
|
1631001600 |
1185 |
 |
Description:
Author: Albert Cheng
Session Name: hg38_ZFP18_1_0_0
Genome Assembly: hg38
Creation Date: 2021-09-07
Views: 1455
|
1631001600 |
1455 |
 |
Description:
Author: JuliaLaz
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2021-09-07
Views: 1309
|
1631001600 |
1309 |
 |
Description:
Author: AbigailLambert
Session Name: Fall19 Yeast test AML
Genome Assembly: sacCer3
Creation Date: 2021-09-12
Views: 1075
|
1631433600 |
1075 |
 |
Description:
Author: layla13
Session Name: Fall21 Yeast test LA
Genome Assembly: sacCer3
Creation Date: 2021-09-05
Views: 1139
|
1630828800 |
1139 |
 |
Description:
Author: gucascau
Session Name: H3-3-2
Genome Assembly: mm10
Creation Date: 2021-09-03
Views: 1147
|
1630656000 |
1147 |
 |
Description:
Author: mallorycunningham
Session Name: Fall21 Yeast Test MC
Genome Assembly: sacCer3
Creation Date: 2021-09-03
Views: 1131
|
1630656000 |
1131 |
 |
Description:
Author: jessicagarth
Session Name: CGEMS yeast JMG test
Genome Assembly: sacCer3
Creation Date: 2021-09-02
Views: 1133
|
1630569600 |
1133 |
 |
Description:
Author: JuliaLaz
Session Name: Fall21 Worm test group#4
Genome Assembly: ce11
Creation Date: 2021-09-02
Views: 883
|
1630569600 |
883 |
 |
Description:
Author: latimeaa
Session Name: CGEMS Worm AAL test
Genome Assembly: ce11
Creation Date: 2021-09-02
Views: 1079
|
1630569600 |
1079 |
 |
Description:
Author: latimeaa
Session Name: Fall21 Worm test group#4
Genome Assembly: ce11
Creation Date: 2021-09-02
Views: 951
|
1630569600 |
951 |
 |
Description:
Author: aazari99
Session Name: CGEMS Worm AA Test
Genome Assembly: ce11
Creation Date: 2021-09-02
Views: 1190
|
1630569600 |
1190 |
 |
Description:
Author: aazari99
Session Name: Fall21 Worm test group#3
Genome Assembly: ce11
Creation Date: 2021-09-02
Views: 912
|
1630569600 |
912 |
 |
Description:
Author: Schne2lk
Session Name: Fall21 Worm test group#3
Genome Assembly: ce11
Creation Date: 2021-09-02
Views: 880
|
1630569600 |
880 |
 |
Description:
Author: laarne
Session Name: CGEMS worm LAN test
Genome Assembly: ce11
Creation Date: 2021-09-02
Views: 1146
|
1630569600 |
1146 |
 |
Description:
Author: whaleycn
Session Name: Fall21 Mouse Test Group(6)
Genome Assembly: mm9
Creation Date: 2021-09-02
Views: 1100
|
1630569600 |
1100 |
 |
Description:
Author: ashleywilkins
Session Name: Fall21 Worm test group #4
Genome Assembly: ce11
Creation Date: 2021-09-02
Views: 980
|
1630569600 |
980 |
 |
Description:
Author: randa2jl
Session Name: Fall21 Yeast Test Group 1
Genome Assembly: sacCer3
Creation Date: 2021-09-02
Views: 1078
|
1630569600 |
1078 |
 |
Description:
Author: asinerbc
Session Name: FAll21 Yeast test group 2
Genome Assembly: sacCer3
Creation Date: 2021-09-02
Views: 1117
|
1630569600 |
1117 |
 |
Description:
Author: AHRImmunity
Session Name: stockingerg_ATACSEQ
Genome Assembly: mm10
Creation Date: 2021-09-01
Views: 1243
|
1630483200 |
1243 |
 |
Description:
Author: AHRImmunity
Session Name: stockingerg_CHIPSEQ
Genome Assembly: mm10
Creation Date: 2021-09-01
Views: 1310
|
1630483200 |
1310 |
 |
Description:
Author: Eldar
Session Name: gene level
Genome Assembly: hg38
Creation Date: 2021-08-27
Views: 1151
|
1630051200 |
1151 |
 |
Description:
Author: aytyuh
Session Name: newnew
Genome Assembly: mm10
Creation Date: 2022-02-16
Views: 896
|
1644998400 |
896 |
 |
Description:
Author: ssdhar
Session Name: hg19 Pim1 Bc
Genome Assembly: hg19
Creation Date: 2021-08-25
Views: 1247
|
1629878400 |
1247 |
 |
Description:
Author: zxtzhangqian
Session Name: Lamprey_WGBS
Genome Assembly: petMar3
Creation Date: 2021-08-25
Views: 1184
|
1629878400 |
1184 |
 |
Description:
Author: schmi4am
Session Name: Fall19 Yeast test AMS
Genome Assembly: sacCer3
Creation Date: 2021-09-05
Views: 1132
|
1630828800 |
1132 |
 |
Description:
Author: ssdhar
Session Name: hg19 mmp11 bc
Genome Assembly: hg19
Creation Date: 2021-08-17
Views: 1204
|
1629187200 |
1204 |
 |
Description:
Author: ssdhar
Session Name: hg19 new H3k27 ac six1
Genome Assembly: hg19
Creation Date: 2021-08-17
Views: 1172
|
1629187200 |
1172 |
 |
Description:
Author: lyj
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2021-08-15
Views: 1140
|
1629014400 |
1140 |
 |
Description:
Author: lyj
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2021-08-15
Views: 1432
|
1629014400 |
1432 |
 |
Description: H3K27ac ChIP-seq count data from A375 melanoma in response to medium-chain fatty acid (octanoate) administration.
Author: thuang316
Session Name: H3K27ac_ChIP_seq_octanoate
Genome Assembly: hg38
Creation Date: 2021-08-15
Views: 945
|
1629014400 |
945 |
 |
Description: ReMap 2022: A database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments Go to <a href="http://remap2022.univ-amu.fr">ReMap2022</a> for more info.
Author: Benoit Ballester
Session Name: US_hg38_ReMap2022
Genome Assembly: hg38
Creation Date: 2021-08-11
Views: 4454
|
1628668800 |
4454 |
 |
Description: ReMap 2022: A database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments Go to <a href="http://remap2022.univ-amu.fr">ReMap2022</a> for more info.
Author: Benoit Ballester
Session Name: US_mm10_ReMap2022
Genome Assembly: mm10
Creation Date: 2021-08-11
Views: 2605
|
1628668800 |
2605 |
 |
Description: ReMap 2022: A database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments Go to <a href="http://remap2022.univ-amu.fr">ReMap2022</a> for more info.
Author: Benoit Ballester
Session Name: US_dm6_ReMap2022
Genome Assembly: dm6
Creation Date: 2021-08-11
Views: 1613
|
1628668800 |
1613 |
 |
Description: ReMap 2022: A database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments Go to <a href="http://remap2022.univ-amu.fr">ReMap2022</a> for more info.
Author: Benoit Ballester
Session Name: US_araTha1_ReMap2022
Genome Assembly: hub_2961057_araTha1
Creation Date: 2021-08-11
Views: 2497
|
1628668800 |
2497 |
 |
Description: This session uses Times font.
Author: brianlee
Session Name: Times_Font
Genome Assembly: hg38
Creation Date: 2021-08-10
Views: 3648
|
1628582400 |
3648 |
 |
Description: A Public Session using the Avant Guard Font.
Author: brianlee
Session Name: AvantG_Font
Genome Assembly: hg19
Creation Date: 2021-08-10
Views: 2637
|
1628582400 |
2637 |
 |
Description: 4931406C07Rik Transcripts
Author: szaman2
Session Name: 4931406C07Rik
Genome Assembly: mm10
Creation Date: 2021-08-10
Views: 1113
|
1628582400 |
1113 |
 |
Description:
Author: jamesli
Session Name: RNA_ATAC_Cerebellum1
Genome Assembly: mm10
Creation Date: 2021-08-06
Views: 943
|
1628236800 |
943 |
 |
Description:
Author: gucascau
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2021-08-05
Views: 1219
|
1628150400 |
1219 |
 |
Description: Sessions shows part of spike protein on SARS-CoV-2 genome. One track shows the reference amino acids for the spike protein. A custom track at the top shows the location of two amino acids that are substituted by prolines to hold the protein in the pre-fusion conformation. A track near the bottom shows the Moderna and Pfizer/BioNtech vaccines with the two prolines substituted for lysine and valine of the reference. The last rack shows current variants of concern (VoC). Reconfigure the vaccine track to see the "different RNA bases" by clicking the gray button to the left of track.
Author: Example
Session Name: wuhCor1_ProPro2
Genome Assembly: wuhCor1
Creation Date: 2021-08-04
Views: 1204
|
1628064000 |
1204 |
 |
Description: The genes HBB and HBD are both expressed in red blood cells, but HBB is also expressed in many other tissues, whereas HBD is expressed in red cells alomst exclusively.
Author: EJ
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2021-08-03
Views: 1225
|
1627977600 |
1225 |
 |
Description:
Author: gonzalezvl
Session Name: pha_v1_ncbi
Genome Assembly: hub_2958085_pha_v1_ncbi
Creation Date: 2021-08-02
Views: 1261
|
1627891200 |
1261 |
 |
Description:
Author: nkfarah
Session Name: snATAC_cerebellum_E12_14
Genome Assembly: mm10
Creation Date: 2021-07-27
Views: 1170
|
1627372800 |
1170 |
 |
Description: The gene HBB and HBD are both expressed in red blood cells, but HBB is also expressed in many other tissues, where as HBD is expressed in red cells almost exclusively.
Author: mw1278
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2021-08-31
Views: 1118
|
1630396800 |
1118 |
 |
Description: This session has Times font selected and italic styling applied, available from the Genome Browser menu -> Configure page (or "c f" shortcut).
Author: PublicSessions
Session Name: Drosophila Duplication with font
Genome Assembly: dm6
Creation Date: 2021-07-21
Views: 1120
|
1626854400 |
1120 |
 |
Description:
Author: jmorlan66
Session Name: PFS_session8
Genome Assembly: hg38
Creation Date: 2021-07-20
Views: 1171
|
1626768000 |
1171 |
 |
Description:
Author: Hanson258
Session Name: H3K27me3_myoblast
Genome Assembly: hg38
Creation Date: 2021-07-16
Views: 1283
|
1626422400 |
1283 |
 |
Description: chıpseq result
Author: hkarakoyun23@ku.edu.tr
Session Name: hazals.
Genome Assembly: hg38
Creation Date: 2024-06-12
Views: 304
|
1718179200 |
304 |
 |
Description:
Author: Jinkil_Jeong
Session Name: Shig_Ad
Genome Assembly: hg19
Creation Date: 2021-07-07
Views: 1182
|
1625644800 |
1182 |
 |
Description:
Author: MatthewMurry
Session Name: danRer11
Genome Assembly: danRer11
Creation Date: 2021-07-07
Views: 1157
|
1625644800 |
1157 |
 |
Description:
Author: smorabito
Session Name: AD_snATAC
Genome Assembly: hg38
Creation Date: 2021-07-07
Views: 1772
|
1625644800 |
1772 |
 |
Description: Human HOX 11 gene mapped on the positive RLFS readings in the bed file.
Author: arazavi1
Session Name: HOX
Genome Assembly: hg19
Creation Date: 2021-07-07
Views: 1198
|
1625644800 |
1198 |
 |
Description: This variant affecting Type 1 collagen is identified as a genetic basis for classical EDS and vascular EDS from this source: https://www.ehlers-danlos.com/eds-types/
This session displays SNV clinically oriented annotation tracks at this locus. A paper describing these tracks and the feature that launches them, "Variant Interpretation: UCSC Genome Browser Recommended Track Sets" is in press (pre-print here: https://www.authorea.com/users/423801/articles/529043-variant-interpretation-ucsc-genome-browser-recommended-track-sets)
Author: Kate
Session Name: Ehlers-Danlos Syndrome COL5A1 variant locus (c.934C>T)
Genome Assembly: hg19
Creation Date: 2021-07-08
Views: 1252
|
1625731200 |
1252 |
 |
Description:
Author: Albert Cheng
Session Name: hg38_JACKIE
Genome Assembly: hg38
Creation Date: 2021-07-01
Views: 1152
|
1625126400 |
1152 |
 |
Description:
Author: Albert Cheng
Session Name: mm10_JACKIE
Genome Assembly: mm10
Creation Date: 2021-07-01
Views: 1174
|
1625126400 |
1174 |
 |
Description: A picture of an assembly hub for Zebu cattle where the current DNA in the region has been sent to the External Tool RSAT Metazoa which discovered a motif, aaacttatagata, within the region. The motif was then highlighted with the Short Match track. The session shows how building assembly hubs enables interoperability with tools beyond the UCSC Genome Browser. To access the External Tools pop-up menu, when browsing a genome hover over the "View" menu and then select External Tools.
Author: PublicSessions
Session Name: ExternalTools
Genome Assembly: hub_2100979_GCF_000247795.1
Creation Date: 2021-07-23
Views: 1330
|
1627027200 |
1330 |
 |
Description:
Author: ashleywilkins
Session Name: hg19 RHO
Genome Assembly: hg19
Creation Date: 2021-09-06
Views: 1145
|
1630915200 |
1145 |
 |
Description:
Author: Schne2lk
Session Name: hg38 RHO
Genome Assembly: hg38
Creation Date: 2021-09-06
Views: 1116
|
1630915200 |
1116 |
 |
Description:
Author: artman1967
Session Name: hg19_IBR finale 061521
Genome Assembly: hg19
Creation Date: 2021-06-15
Views: 1204
|
1623744000 |
1204 |
 |
Description:
Author: Shevelyov
Session Name: LaminDamID in SpG and SpCs
Genome Assembly: dm3
Creation Date: 2021-06-12
Views: 1281
|
1623484800 |
1281 |
 |
Description:
Author: PolinaKananykina
Session Name: polina_kananykina
Genome Assembly: hg19
Creation Date: 2021-06-09
Views: 1152
|
1623225600 |
1152 |
 |
Description:
Author: labashmanov
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2021-06-09
Views: 1252
|
1623225600 |
1252 |
 |
Description:
Author: PolinaKananykina
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2021-06-09
Views: 1148
|
1623225600 |
1148 |
 |
Description:
Author: art591
Session Name: hse21_H3K27ac_MCF-7_human
Genome Assembly: hg19
Creation Date: 2021-06-09
Views: 1147
|
1623225600 |
1147 |
 |
Description: THRB
Author: Ruffi2ec
Session Name: Ruffing_ Lab 4-10 THRB
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 352
|
1712736000 |
352 |
 |
Description: PDE6A
Author: Ruffi2ec
Session Name: Ruffing_ Lab 4-10 PDE6A
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 373
|
1712736000 |
373 |
 |
Description: RHO edit
Author: Ruffi2ec
Session Name: Ruffing_ Lab 4-10 RHO
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 342
|
1712736000 |
342 |
 |
Description:
Author: xiaolinwei123
Session Name: hg38_gata6
Genome Assembly: hg38
Creation Date: 2021-05-26
Views: 1118
|
1622016000 |
1118 |
 |
Description:
Author: Xenia
Session Name: DNA methilarion 14chr
Genome Assembly: mm10
Creation Date: 2021-05-24
Views: 1186
|
1621843200 |
1186 |
 |
Description:
Author: gemtang
Session Name: histone
Genome Assembly: hg38
Creation Date: 2021-06-04
Views: 1167
|
1622793600 |
1167 |
 |
Description:
Author: Maksimov_Denis
Session Name: RNA_seq
Genome Assembly: mm10
Creation Date: 2021-05-24
Views: 1257
|
1621843200 |
1257 |
 |
Description: Regions on GRCh37 where discordant exome variant calls are enriched when mapped on GRCh38.
Author: mdawood
Session Name: DISCREPS_GRCh37
Genome Assembly: hg19
Creation Date: 2021-05-17
Views: 1643
|
1621238400 |
1643 |
 |
Description: Regions on GRCh38 where discordant exome variant calls are enriched when mapped on GRCh37.
Author: mdawood
Session Name: DISCREPs_GRCh38
Genome Assembly: hg38
Creation Date: 2021-05-17
Views: 1387
|
1621238400 |
1387 |
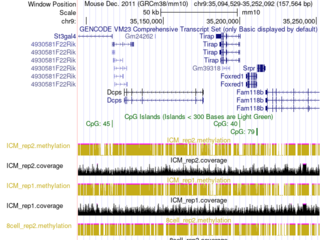 |
Description:
Author: jojobajo
Session Name: SarkisyanA
Genome Assembly: mm10
Creation Date: 2021-05-17
Views: 1224
|
1621238400 |
1224 |
 |
Description:
Author: manu90
Session Name: paciente2_trasloca
Genome Assembly: hg19
Creation Date: 2021-05-12
Views: 1241
|
1620806400 |
1241 |
 |
Description:
Author: manu90
Session Name: ZMIZ
Genome Assembly: hg19
Creation Date: 2021-05-12
Views: 1469
|
1620806400 |
1469 |
 |
Description:
Author: Maksimov_Denis
Session Name: chromatine states
Genome Assembly: hg19
Creation Date: 2021-05-11
Views: 1209
|
1620720000 |
1209 |
 |
Description: tRNA2
Author: ck-j
Session Name: tRNA2
Genome Assembly: hg38
Creation Date: 2022-12-27
Views: 732
|
1672128000 |
732 |
 |
Description:
Author: eva re
Session Name: mm10_er
Genome Assembly: mm10
Creation Date: 2021-05-10
Views: 1151
|
1620633600 |
1151 |
 |
Description:
Author: eva re
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2021-05-10
Views: 1145
|
1620633600 |
1145 |
 |
Description:
Author: jdpachec
Session Name: Pacheco-PS2-ACE2-session
Genome Assembly: hg19
Creation Date: 2021-05-01
Views: 1195
|
1619856000 |
1195 |
 |
Description:
Author: amatsiev
Session Name: Matsiev-PS2-ACE2-session
Genome Assembly: hg19
Creation Date: 2021-04-30
Views: 1217
|
1619769600 |
1217 |
 |
Description:
Author: kailasdhond
Session Name: BME 110 PS 2
Genome Assembly: hg19
Creation Date: 2021-04-30
Views: 1511
|
1619769600 |
1511 |
 |
Description:
Author: mbule
Session Name: Bule-PS2-ACE2-session
Genome Assembly: hg19
Creation Date: 2021-04-30
Views: 1237
|
1619769600 |
1237 |
 |
Description:
Author: mai.baker
Session Name: hg38_SAT1_bed
Genome Assembly: hg38
Creation Date: 2021-04-28
Views: 1208
|
1619596800 |
1208 |
 |
Description:
Author: beoungle
Session Name: hg19 enhancer and infos
Genome Assembly: hg19
Creation Date: 2021-04-26
Views: 1963
|
1619424000 |
1963 |
 |
Description:
Author: ramace
Session Name: Mace--PS2-ACE2-session
Genome Assembly: hg19
Creation Date: 2021-04-25
Views: 1146
|
1619337600 |
1146 |
 |
Description:
Author: Maksimov_Denis
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2021-05-11
Views: 1177
|
1620720000 |
1177 |
 |
Description:
Author: abenet
Session Name: HumanMutationFig2
Genome Assembly: hg19
Creation Date: 2021-05-19
Views: 1286
|
1621411200 |
1286 |
 |
Description:
Author: abenet
Session Name: HumanMutationFig4
Genome Assembly: hg19
Creation Date: 2021-05-19
Views: 1242
|
1621411200 |
1242 |
 |
Description:
Author: jojshelt
Session Name: ACE 2 04/17/21
Genome Assembly: hg38
Creation Date: 2021-04-17
Views: 1166
|
1618646400 |
1166 |
 |
Description:
Author: jojshelt
Session Name: SARS CoV 2 04/15/21
Genome Assembly: wuhCor1
Creation Date: 2021-04-15
Views: 3387
|
1618473600 |
3387 |
 |
Description:
Author: gatupovmikhail
Session Name: mm10_hw1
Genome Assembly: mm10
Creation Date: 2021-04-14
Views: 2988
|
1618387200 |
2988 |
 |
Description:
Author: Maksimov_Denis
Session Name: bisulfite sequencing
Genome Assembly: mm10
Creation Date: 2021-04-14
Views: 1176
|
1618387200 |
1176 |
 |
Description: Public session with the basic tracks to identify putative endothelial enhancers, as described in the published method chapter:
Neal A, Rodriguez-Caro H, De Val S. Finding and Verifying Enhancers for Endothelial-Expressed Genes. Methods Mol Biol. 2022;2441:351-368. doi: 10.1007/978-1-0716-2059-5_28. PMID: 35099751.
If used, please cite the publication.
Author: Helena Rodriguez-Caro
Author: HRC
Session Name: AngiogenesisProtocol_2021_DeVal
Genome Assembly: hg19
Creation Date: 2021-04-09
Views: 1246
|
1617955200 |
1246 |
 |
Description:
Author: qscmki
Session Name: WT-RNA-ribo-seq
Genome Assembly: mm10
Creation Date: 2021-04-08
Views: 1308
|
1617868800 |
1308 |
 |
Description:
Author: gbenegas
Session Name: tabulamuris
Genome Assembly: mm10
Creation Date: 2021-04-06
Views: 1394
|
1617696000 |
1394 |
 |
Description:
Author: gbenegas
Session Name: primarymotorcortex
Genome Assembly: mm10
Creation Date: 2021-04-06
Views: 1360
|
1617696000 |
1360 |
 |
Description:
Author: JFormosa
Session Name: Hwk6Pt3
Genome Assembly: hg19
Creation Date: 2022-02-22
Views: 873
|
1645516800 |
873 |
 |
Description: This session shows a heat map display using bigBed tracks. A new bigHeat python program helps build a heat-map style display of tracks in various colors. It takes an input BED file and an expression-matrix and generates one bigBed file per column, colored by the values in the matrix. For SARS-2 there are assays where peptides are spotted onto a microarray and antibodies extracted from blood are run over the array and the bigHeat tool was created to aid in visualizing this microarray type data into a collection of stacked tracks in the Browser seen in this session.
Author: brianlee
Session Name: HeatMap
Genome Assembly: wuhCor1
Creation Date: 2021-04-06
Views: 1203
|
1617696000 |
1203 |
 |
Description:
Author: ldebiasio
Session Name: DeBiaso-PS2-ACE2-session
Genome Assembly: hg19
Creation Date: 2021-04-27
Views: 1200
|
1619510400 |
1200 |
 |
Description:
Author: marijazzz
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2021-03-28
Views: 1303
|
1616918400 |
1303 |
 |
Description:
Author: asalisbury1
Session Name: mm9999
Genome Assembly: mm9
Creation Date: 2021-03-25
Views: 1274
|
1616659200 |
1274 |
 |
Description:
Author: vrlopez
Session Name: Spring 22 Yeast test VL
Genome Assembly: sacCer3
Creation Date: 2022-01-28
Views: 891
|
1643356800 |
891 |
 |
Description:
Author: Cornelia
Session Name: danRer10_pEGFP-N1_RNA
Genome Assembly: danRer10
Creation Date: 2021-03-04
Views: 1254
|
1614844800 |
1254 |
 |
Description:
Author: butlertj3
Session Name: sars_cov2_g4_stem-loops
Genome Assembly: wuhCor1
Creation Date: 2020-11-26
Views: 1496
|
1606377600 |
1496 |
 |
Description: This session shows ChIP-on-ChEP-seq data from Macaca fascicularis testis for H3K27Ac, REC8, RAD21L, SMC1B, STAG3, CTCF, and BORIS/CTCFL. Data in this session are from the PNAS paper:
A. Boukaba, J. Liu, C. Ward, Q. Wu1, A. Arnaoutov, J. Liang1, E. M. Pugacheva, M. Dasso, V. Lobanenkov, M. Esteban, and A. V. Strunnikov (2022) Ectopic expression of meiotic cohesin generates chromosome instability in cancer cell line. Proc Natl Acad Sci U S A. Oct 4;119(40):e2204071119. doi: 10.1073/pnas.2204071119.
Author: Strunnik
Session Name: cohesins_CTCF_BORIS_K27Ac_macFas5_pub
Genome Assembly: macFas5
Creation Date: 2021-02-28
Views: 985
|
1614499200 |
985 |
 |
Description:
Author: jhj981111
Session Name: Pde4b
Genome Assembly: mm10
Creation Date: 2021-03-02
Views: 1239
|
1614672000 |
1239 |
 |
Description:
Author: Mohamed Hisham
Session Name: hg19-HBB
Genome Assembly: hg19
Creation Date: 2021-03-02
Views: 1198
|
1614672000 |
1198 |
 |
Description:
Author: veronicaburns
Session Name: mm10_210225
Genome Assembly: mm10
Creation Date: 2021-02-25
Views: 1440
|
1614240000 |
1440 |
 |
Description:
Author: abenet
Session Name: HumanMutationFig3
Genome Assembly: hg19
Creation Date: 2021-05-19
Views: 1191
|
1621411200 |
1191 |
 |
Description:
Author: elencampoy
Session Name: HsInv0124
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1365
|
1614153600 |
1365 |
 |
Description:
Author: elencampoy
Session Name: HsInv1057
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1438
|
1614153600 |
1438 |
 |
Description:
Author: elencampoy
Session Name: HsInv1051
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1419
|
1614153600 |
1419 |
 |
Description:
Author: elencampoy
Session Name: HsInv0830
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1193
|
1614153600 |
1193 |
 |
Description:
Author: elencampoy
Session Name: HsInv0403
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1374
|
1614153600 |
1374 |
 |
Description:
Author: elencampoy
Session Name: HsInv0397
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1244
|
1614153600 |
1244 |
 |
Description:
Author: elencampoy
Session Name: HsInv0395
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1193
|
1614153600 |
1193 |
 |
Description:
Author: elencampoy
Session Name: HsInv0393
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1196
|
1614153600 |
1196 |
 |
Description:
Author: elencampoy
Session Name: HsInv0390
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1272
|
1614153600 |
1272 |
 |
Description:
Author: elencampoy
Session Name: hsinv0389
Genome Assembly: hg19
Creation Date: 2021-02-24
Views: 1200
|
1614153600 |
1200 |
 |
Description:
Author: elencampoy
Session Name: HsInv0370
Genome Assembly: hg19
Creation Date: 2021-02-23
Views: 1240
|
1614067200 |
1240 |
 |
Description:
Author: elencampoy
Session Name: HsInv1124
Genome Assembly: hg19
Creation Date: 2021-02-23
Views: 1250
|
1614067200 |
1250 |
 |
Description:
Author: elencampoy
Session Name: HsInv0290
Genome Assembly: hg19
Creation Date: 2021-02-23
Views: 1188
|
1614067200 |
1188 |
 |
Description:
Author: arthurcheng
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2021-02-20
Views: 1214
|
1613808000 |
1214 |
 |
Description: Genome-wide profiling of histone H1 variants and H3K9me3 in T47D-MTVL cells upon multiple H1 depletion.
Author: NSP
Session Name: GSE156036_GSE166645
Genome Assembly: hg19
Creation Date: 2020-09-08
Views: 1559
|
1599552000 |
1559 |
 |
Description:
Author: elencampoy
Session Name: HsInv0786
Genome Assembly: hg19
Creation Date: 2021-02-18
Views: 1234
|
1613635200 |
1234 |
 |
Description:
Author: elencampoy
Session Name: HsInv0344
Genome Assembly: hg19
Creation Date: 2021-02-18
Views: 1192
|
1613635200 |
1192 |
 |
Description:
Author: elencampoy
Session Name: HsInv0340
Genome Assembly: hg19
Creation Date: 2021-02-23
Views: 1234
|
1614067200 |
1234 |
 |
Description:
Author: elencampoy
Session Name: HsInv0228
Genome Assembly: hg19
Creation Date: 2021-02-23
Views: 1193
|
1614067200 |
1193 |
 |
Description:
Author: elencampoy
Session Name: HsInv0396
Genome Assembly: hg19
Creation Date: 2021-02-17
Views: 1222
|
1613548800 |
1222 |
 |
Description:
Author: elencampoy
Session Name: hsinv0097
Genome Assembly: hg19
Creation Date: 2021-02-17
Views: 1218
|
1613548800 |
1218 |
 |
Description: Posterior Column Ataxia with Retinitis Pigmentosa (PCARP) is a genetic disorder caused by mutations in the FLVCR1 gene that codes for Feline Leukemia Virus Subgroup C Cellular Receptor 1. Mutations in this heme transport protein, can lead to heme toxicity which affects vision and the nervous system. Symptoms begin in childhood and can include Tunnel-like visual loss, night blindness, and photophobia. Through adulthood sensory function decreases, and symptoms of atrophy, loss of touch, and scoliosis, are common. There are four variants of the FLVCR1 gene that cause PCARP. Puffenburger et. al identified a novel variant at position 361 of the FLCR1 gene, an Asparagine-to-Aspartic Acid, missense mutation which is highlighted blue. The Puffenburger variant (blue) is the current focus of the track, if you zoom to see the whole FLVCR gene you will see there are three other variants designated by the OMIM Alleles Track. Two additional variants are also located in exon 1, and a third in exon 8. At position 574 there is a variant resulting in a cysteine-to-arginine missense mutation which is highlighted green, and at position 721, an alanine-to-threonine missense mutation which is highlighted yellow. In Exon 8 at position 1477 there is a variant resulting in a glycine-to-arginine missense mutation. The Clinvar Variants track classifies all four as pathogenic missense mutations. Pathogenic and Likely Pathogenic variants are colored red, benign variants are colored green, and variants of uncertain significance are colored gray. The Conservation track shows that the regions where all four variants are highly conserved across several species. There is no amino acid variation between chimps, dogs, cattle, cattle, mice, fowl, and zebrafish. Conservation across all of these species indicates these regions are important to vertebrates, and explains why these missense mutations are damaging.
Author: jrhemm
Session Name: Juniata Bioinformatics JHNH
Genome Assembly: hg19
Creation Date: 2021-02-11
Views: 1242
|
1613030400 |
1242 |
 |
Description: This session shows bosTau9 for a gene also found in Human_PIK3CG which was described in a PAG 2020 meeting.
Author: brianlee
Session Name: bosTau9_Human_PIK3CG
Genome Assembly: bosTau9
Creation Date: 2021-02-12
Views: 1208
|
1613116800 |
1208 |
 |
Description:
Author: soniagoozee
Session Name: bushbaby
Genome Assembly: hg38
Creation Date: 2021-02-11
Views: 1144
|
1613030400 |
1144 |
 |
Description:
Author: elencampoy
Session Name: inversions
Genome Assembly: hg19
Creation Date: 2021-02-11
Views: 1201
|
1613030400 |
1201 |
 |
Description:
Author: raceschaeffer
Session Name: mm10_task2_int
Genome Assembly: mm10
Creation Date: 2021-02-10
Views: 1230
|
1612944000 |
1230 |
 |
Description:
Author: raceschaeffer
Session Name: mm10_task1
Genome Assembly: mm10
Creation Date: 2021-02-10
Views: 1608
|
1612944000 |
1608 |
 |
Description:
Author: joel_mundkl
Session Name: Alg 13 w/ tracks
Genome Assembly: mm10
Creation Date: 2021-02-10
Views: 1265
|
1612944000 |
1265 |
 |
Description:
Author: joel_mundkl
Session Name: All 3 tracks session
Genome Assembly: mm10
Creation Date: 2021-02-10
Views: 1184
|
1612944000 |
1184 |
 |
Description:
Author: joel_mundkl
Session Name: Gli3 and HH tracks
Genome Assembly: mm10
Creation Date: 2021-02-10
Views: 1406
|
1612944000 |
1406 |
 |
Description:
Author: joel_mundkl
Session Name: Gli3 limb binding mm10 (ptch1)
Genome Assembly: mm10
Creation Date: 2021-02-10
Views: 1205
|
1612944000 |
1205 |
 |
Description:
Author: qhernand
Session Name: Hernandez-PS2-ACE2-session
Genome Assembly: hg19
Creation Date: 2021-04-30
Views: 1420
|
1619769600 |
1420 |
 |
Description:
Author: alecmilazzo
Session Name: mm10_1
Genome Assembly: mm10
Creation Date: 2021-02-09
Views: 1665
|
1612857600 |
1665 |
 |
Description:
Author: as3838
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2021-02-08
Views: 1271
|
1612771200 |
1271 |
 |
Description:
Author: PaoloM1990
Session Name: ACTB_qPCR primers
Genome Assembly: hg19
Creation Date: 2021-02-02
Views: 1617
|
1612252800 |
1617 |
 |
Description:
Author: mdrcetin
Session Name: hg38_encode_bcell_cd34myeloidprogenitor_naturalkiller
Genome Assembly: hg38
Creation Date: 2021-11-16
Views: 933
|
1637049600 |
933 |
 |
Description:
Author: mdrcetin
Session Name: hg38_encode_ucsf4_hyyees64_h1_h9
Genome Assembly: hg38
Creation Date: 2021-11-16
Views: 1450
|
1637049600 |
1450 |
 |
Description:
Author: mdrcetin
Session Name: hg38_k562_encode_minus_strand_only
Genome Assembly: hg38
Creation Date: 2021-11-16
Views: 937
|
1637049600 |
937 |
 |
Description:
Author: molcov
Session Name: Human Duplication
Genome Assembly: hg19
Creation Date: 2021-01-31
Views: 1270
|
1612080000 |
1270 |
 |
Description: coronavirus spike protein region with some relevant tracks turned on, including variants of concern (VoC).
Author: Example
Session Name: wuhCor1_spike
Genome Assembly: wuhCor1
Creation Date: 2021-01-25
Views: 3609
|
1611561600 |
3609 |
 |
Description:
Author: Dongyao_Liu
Session Name: mm9_BAML1_and_CLOCK
Genome Assembly: mm9
Creation Date: 2021-04-13
Views: 1180
|
1618300800 |
1180 |
 |
Description: The hub shows association of copy number changes of VNTRs (Variable Number Tander Repeat) with local gene expression and CpG methylation in two discovery cohorts ane one replication cohort for which PCR-free Illumina WGS data were available.<br>
Read more here: https://gargp01.u.hpc.mssm.edu/ucsc_exp_meth_vntr.html
Author: brianlee
Session Name: VNTR
Genome Assembly: hg19
Creation Date: 2021-01-22
Views: 1264
|
1611302400 |
1264 |
 |
Description:
Author: elencampoy
Session Name: Hsinv0209
Genome Assembly: hg19
Creation Date: 2021-02-23
Views: 1225
|
1614067200 |
1225 |
 |
Description:
Author: elencampoy
Session Name: HsInv0030
Genome Assembly: hg19
Creation Date: 2021-02-23
Views: 1210
|
1614067200 |
1210 |
 |
Description:
Author: BOPOHDR
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2021-01-12
Views: 1713
|
1610438400 |
1713 |
 |
Description:
Author: ashleywilkins
Session Name: hg19 RHO 2
Genome Assembly: hg19
Creation Date: 2021-09-06
Views: 1144
|
1630915200 |
1144 |
 |
Description: CAG repeats found in the RUNX2 gene. Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: anom_fig9
Genome Assembly: hg19
Creation Date: 2022-08-19
Views: 843
|
1660896000 |
843 |
 |
Description: Zoom in on CAG repeats found in the gene MED12. Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: anom_fig7
Genome Assembly: hg19
Creation Date: 2022-08-19
Views: 849
|
1660896000 |
849 |
 |
Description: Two different areas of CAG repeats found in the MED12 gene. Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: anom_fig6
Genome Assembly: hg19
Creation Date: 2022-08-19
Views: 956
|
1660896000 |
956 |
 |
Description:
Author: as3838
Session Name: PATRICKDATA
Genome Assembly: hg19
Creation Date: 2021-02-08
Views: 1312
|
1612771200 |
1312 |
 |
Description:
Author: AnnaClaireR1
Session Name: mm10_3tracks
Genome Assembly: mm10
Creation Date: 2021-02-01
Views: 1497
|
1612166400 |
1497 |
 |
Description:
Author: AnnaClaireR1
Session Name: mm10_testupload
Genome Assembly: mm10
Creation Date: 2021-02-01
Views: 1278
|
1612166400 |
1278 |
 |
Description:
Author: PaoloM1990
Session Name: GAPDH_qPCR primers
Genome Assembly: hg19
Creation Date: 2021-02-02
Views: 1258
|
1612252800 |
1258 |
 |
Description:
Author: PaoloM1990
Session Name: ATP6V0E1_qPCR primers
Genome Assembly: hg19
Creation Date: 2021-02-02
Views: 1199
|
1612252800 |
1199 |
 |
Description:
Author: PaoloM1990
Session Name: GBA_qPCR primers
Genome Assembly: hg19
Creation Date: 2021-02-02
Views: 1300
|
1612252800 |
1300 |
 |
Description:
Author: PaoloM1990
Session Name: CTSD_qPCR primers
Genome Assembly: hg19
Creation Date: 2021-02-02
Views: 1169
|
1612252800 |
1169 |
 |
Description:
Author: PaoloM1990
Session Name: CTSB_qPCR primers
Genome Assembly: hg19
Creation Date: 2021-02-02
Views: 1301
|
1612252800 |
1301 |
 |
Description:
Author: PaoloM1990
Session Name: CTSL_qPCR primers
Genome Assembly: hg19
Creation Date: 2021-02-02
Views: 1224
|
1612252800 |
1224 |
 |
Description:
Author: PaoloM1990
Session Name: MCOLN1_qPCR primers
Genome Assembly: hg19
Creation Date: 2021-02-02
Views: 1370
|
1612252800 |
1370 |
 |
Description:
Author: AnnaClaireR1
Session Name: mm10_smoc
Genome Assembly: mm10
Creation Date: 2021-02-02
Views: 1540
|
1612252800 |
1540 |
 |
Description: This session helps demonstrate the Simple Tandem Repeats by TRF track. It includes a Short Match track that is simply highlighting the sequence <b>ctctcgct</b> within the window and this reflects how a programmatic approach can annotate the genome, where the Tandem Repeats Finder (TRF) program helps discover and annotate possibly imperfect repeats as well as exact matches. Users who are interested in STRs may want to look at a resource called STRBase https://strbase.nist.gov/index.htm which was founded in 2001 and includes a Variant Allele Reports page for those searching for allele or frequency count information.
Author: brianlee
Session Name: hg38_STR
Genome Assembly: hg38
Creation Date: 2021-01-07
Views: 1582
|
1610006400 |
1582 |
 |
Description: This session shows an assembly hub on mink that can be loaded with this quick link: https://genome.ucsc.edu/h/GCA_900108605.1<br>
The link can also be searched with the position= term: https://genome.ucsc.edu/h/GCA_900108605.1?position=ACE2 <br>
It can also have custom data displayed on it, in this case an interact file that can be loaded under My Data and Custom Tracks.
Author: brianlee
Session Name: Mink_hub_interact
Genome Assembly: hub_2637617_GCA_900108605.1
Creation Date: 2021-01-07
Views: 1527
|
1610006400 |
1527 |
 |
Description:
Author: sarahjc
Session Name: FM GR ChIP Miseq
Genome Assembly: hg38
Creation Date: 2021-01-02
Views: 1427
|
1609574400 |
1427 |
 |
Description:
Author: BOPOHDR
Session Name: GRCh37_CNV_SNV_2
Genome Assembly: hg19
Creation Date: 2020-12-21
Views: 1415
|
1608537600 |
1415 |
 |
Description:
Author: Libby3121
Session Name: mm39 FoxA2 sites
Genome Assembly: mm39
Creation Date: 2020-12-20
Views: 1344
|
1608451200 |
1344 |
 |
Description:
Author: BOPOHDR
Session Name: GRCh37_CNV_SNV
Genome Assembly: hg19
Creation Date: 2020-12-19
Views: 1922
|
1608364800 |
1922 |
 |
Description:
Author: solbru
Session Name: hg19_TANGO6
Genome Assembly: hg19
Creation Date: 2020-12-18
Views: 1518
|
1608278400 |
1518 |
 |
Description:
Author: molcov
Session Name: Drosophila Duplication
Genome Assembly: dm6
Creation Date: 2021-01-30
Views: 1298
|
1611993600 |
1298 |
 |
Description:
Author: joliemb
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2021-02-04
Views: 1279
|
1612425600 |
1279 |
 |
Description:
Author: mongerc
Session Name: mm10BMI1
Genome Assembly: mm10
Creation Date: 2020-12-16
Views: 1474
|
1608105600 |
1474 |
 |
Description:
Author: rpe13002
Session Name: hg38_CT83_iso3
Genome Assembly: hg38
Creation Date: 2020-12-13
Views: 1571
|
1607846400 |
1571 |
 |
Description:
Author: rpe13002
Session Name: hg38_TMPRSS4_iso4
Genome Assembly: hg38
Creation Date: 2020-12-12
Views: 1430
|
1607760000 |
1430 |
 |
Description:
Author: rpe13002
Session Name: hg38_CT83
Genome Assembly: hg38
Creation Date: 2020-12-12
Views: 1404
|
1607760000 |
1404 |
 |
Description:
Author: rpe13002
Session Name: hg38_ADAM9_iso27
Genome Assembly: hg38
Creation Date: 2020-12-12
Views: 1440
|
1607760000 |
1440 |
 |
Description:
Author: rpe13002
Session Name: hg38_ITGA6_iso20
Genome Assembly: hg38
Creation Date: 2020-12-12
Views: 1430
|
1607760000 |
1430 |
 |
Description:
Author: mongerc
Session Name: Brien2020AllTracks
Genome Assembly: hg38
Creation Date: 2020-12-07
Views: 1477
|
1607328000 |
1477 |
 |
Description:
Author: mongerc
Session Name: Brien2020ChIP-RX
Genome Assembly: hg38
Creation Date: 2020-12-07
Views: 1432
|
1607328000 |
1432 |
 |
Description:
Author: JZhaoARUP
Session Name: hg19_ARUP_CMA_multi_gene
Genome Assembly: hg19
Creation Date: 2020-11-14
Views: 2255
|
1605340800 |
2255 |
 |
Description:
Author: JZhaoARUP
Session Name: hg19_ARUP_CMA_single_gene
Genome Assembly: hg19
Creation Date: 2020-11-14
Views: 1812
|
1605340800 |
1812 |
 |
Description:
Author: sarahjc
Session Name: Nextseq Dedup ESF BAC SS 113020
Genome Assembly: hg38
Creation Date: 2020-11-30
Views: 1404
|
1606723200 |
1404 |
 |
Description:
Author: ustiugova_al
Session Name: au
Genome Assembly: hg38
Creation Date: 2020-11-27
Views: 1446
|
1606464000 |
1446 |
 |
Description: Shown in Fig 1 of "Using data from the 100,000 Genomes Project to resolve conflicting interpretations of a recurrent TUBB2A mutation" (PMID: 33547136).
Shows TUBB2A and positions of de novo variants seen in the 100kGP. Also shown are cismorphisms (red) that differ between TUBB2A, TUBB2B and TUBB4A, TUBB3, TUBB6 and the TUBB2BP1 pseudogene. Primer positions used for variant validation are also plotted.
Author: AlistairP
Session Name: TUBB2A_v5
Genome Assembly: hg38
Creation Date: 2020-11-26
Views: 329
|
1606377600 |
329 |
 |
Description:
Author: ck-j
Session Name: RNAseq-WT/KO-chr12:113,418,558-113,422,730
Genome Assembly: mm10
Creation Date: 2020-11-21
Views: 954
|
1605945600 |
954 |
 |
Description:
Author: ck-j
Session Name: PCPB1-chr12:113,418,558-113,422,730
Genome Assembly: mm10
Creation Date: 2020-11-17
Views: 971
|
1605600000 |
971 |
 |
Description: KJ8Vehicle = 0d +TGFB
KJ15eEMT = 2d +TGFB
KJ12pEMT = 4d +TGFB
KJ14Trans = 4d +TGFB followed by 4d TGFB withdrawal
KJ11fMET2 = 4d +TGFB followed by 10d TGFB withdrawal
KJ9fEMT = 10d +TGFB
KJ13pMET = 10d +TGFB followed by 4d TGFB withdrawal
KJ10fMET1 = 10d +TGFB followed by 10d TGFB withdrawal
Author: kelsey_johnson1
Session Name: Reversible EMT ATAC-seq peaks
Genome Assembly: hg19
Creation Date: 2020-11-20
Views: 1337
|
1605859200 |
1337 |
 |
Description: NRA-Anterograde Regulation of Nuclear-encoded Mitochondrial Genes and FGF21 Signaling by Hepatic Histone Demethylase LSD1
Author: qiushi
Session Name: RNAseq
Genome Assembly: mm10
Creation Date: 2020-11-12
Views: 1482
|
1605168000 |
1482 |
 |
Description: NRA-Anterograde Regulation of Nuclear-encoded Mitochondrial Genes and FGF21 Signaling by Hepatic Histone Demethylase LSD1
Author: qiushi
Session Name: ChIPseq
Genome Assembly: mm10
Creation Date: 2020-11-12
Views: 1459
|
1605168000 |
1459 |
 |
Description:
Author: kkaczor
Session Name: cirm_ucla_2
Genome Assembly: hg38
Creation Date: 2020-11-11
Views: 1534
|
1605081600 |
1534 |
 |
Description:
Author: jranish
Session Name: Castrillo LXR ChIP
Genome Assembly: mm10
Creation Date: 2020-10-27
Views: 1176
|
1603785600 |
1176 |
 |
Description:
Author: zgtman
Session Name: Genescan_3_hg38_SRY
Genome Assembly: hg38
Creation Date: 2021-01-12
Views: 1667
|
1610438400 |
1667 |
 |
Description:
Author: manu90
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2020-11-15
Views: 1189
|
1605427200 |
1189 |
 |
Description:
Author: evechen97
Session Name: EC-39545454-MEDG580-UBC-2020
Genome Assembly: hg38
Creation Date: 2021-10-10
Views: 1144
|
1633852800 |
1144 |
 |
Description:
Author: aurbanow
Session Name: tammer
Genome Assembly: mm10
Creation Date: 2020-11-25
Views: 1469
|
1606291200 |
1469 |
 |
Description:
Author: rafal.czapiewski@ed.ac.uk
Session Name: BAC_FOSMID
Genome Assembly: mm9
Creation Date: 2020-10-16
Views: 1695
|
1602835200 |
1695 |
 |
Description:
Author: psharma
Session Name: 6. PS-75227116-MEDG580-UBC-2020
Genome Assembly: hg38
Creation Date: 2020-10-06
Views: 1376
|
1601971200 |
1376 |
 |
Description:
Author: williamhconrad
Session Name: TSS-evaluation-lite-2
Genome Assembly: hg19
Creation Date: 2021-10-07
Views: 1055
|
1633593600 |
1055 |
 |
Description:
Author: williamhconrad
Session Name: TSS-evaluation-lite
Genome Assembly: hg19
Creation Date: 2021-10-07
Views: 1065
|
1633593600 |
1065 |
 |
Description:
Author: fushenchu
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2020-09-28
Views: 1489
|
1601280000 |
1489 |
 |
Description:
Author: federikr
Session Name: CRX
Genome Assembly: hg38
Creation Date: 2020-09-28
Views: 1484
|
1601280000 |
1484 |
 |
Description: ClinGen tracks show dosage sensitivity (deletion or gains will cause disease or not) of described copy number region (ISCA) and single genes in the region and curation of association of diseases to the genes in these region.
Author: abenet
Session Name: clinical-ClinGenCuration
Genome Assembly: hg19
Creation Date: 2020-09-25
Views: 1492
|
1601020800 |
1492 |
 |
Description:
Author: zahra jafari
Session Name: homwork
Genome Assembly: hg19
Creation Date: 2021-12-03
Views: 905
|
1638518400 |
905 |
 |
Description: This session shows a portion of the ALDH2 gene, one of the two genes associated with alcohol intolerance in East Asian populations, in its entirety. The "ClinVar" track is enabled and shows common nucleotide variants in the region. Clicking into a rsID provides more information regarding the phenotypic, and possibly pathogenic, consequences of a variant. Highlighted is rs671, which is the variant that causes “alcohol flush syndrome”; This condition is seen in approximately 30% to 50% of East Asian individuals.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_ALDH2clinvar
Genome Assembly: hg19
Creation Date: 2022-07-21
Views: 845
|
1658390400 |
845 |
 |
Description:
Author: williamhconrad
Session Name: TSS-evaluation
Genome Assembly: hg19
Creation Date: 2020-09-21
Views: 2113
|
1600675200 |
2113 |
 |
Description:
Author: kianayaghoobian
Session Name: TAS2R38 UniProt
Genome Assembly: hg19
Creation Date: 2020-09-20
Views: 1461
|
1600588800 |
1461 |
 |
Description:
Author: kianayaghoobian
Session Name: TAS2R38 group1
Genome Assembly: hg19
Creation Date: 2020-09-20
Views: 1838
|
1600588800 |
1838 |
 |
Description:
Author: jzhicks07
Session Name: TAS2R38 JMH_1
Genome Assembly: hg19
Creation Date: 2020-09-14
Views: 1523
|
1600070400 |
1523 |
 |
Description:
Author: manu90
Session Name: TAD!!
Genome Assembly: hg19
Creation Date: 2021-06-03
Views: 1176
|
1622707200 |
1176 |
 |
Description:
Author: jzhicks07
Session Name: TAS2R38 JMH
Genome Assembly: hg19
Creation Date: 2020-09-12
Views: 1521
|
1599897600 |
1521 |
 |
Description:
Author: zgtman
Session Name: OTOA
Genome Assembly: hg38
Creation Date: 2021-10-11
Views: 1111
|
1633939200 |
1111 |
 |
Description:
Author: rstarks23
Session Name: e9.5Placenta_HistoneModification
Genome Assembly: mm10
Creation Date: 2020-09-09
Views: 1450
|
1599638400 |
1450 |
 |
Description:
Author: AMOza
Session Name: LMM_Conservation
Genome Assembly: hg19
Creation Date: 2020-09-09
Views: 1553
|
1599638400 |
1553 |
 |
Description:
Author: maddieevans9
Session Name: TAS2R38 group(ME,BZ,CB)
Genome Assembly: hg19
Creation Date: 2020-09-09
Views: 2189
|
1599638400 |
2189 |
 |
Description:
Author: gage2ea
Session Name: Fall20 Worm test group 3
Genome Assembly: ce11
Creation Date: 2020-09-08
Views: 1531
|
1599552000 |
1531 |
 |
Description:
Author: sconyejekwe
Session Name: Fall20 Mouse test group#5
Genome Assembly: mm9
Creation Date: 2020-09-08
Views: 1256
|
1599552000 |
1256 |
 |
Description:
Author: calvinc98
Session Name: Fall20 Mouse test group 6
Genome Assembly: mm9
Creation Date: 2020-09-08
Views: 1467
|
1599552000 |
1467 |
 |
Description:
Author: occamslaser
Session Name: Fall20 Worm test group# 4
Genome Assembly: ce11
Creation Date: 2020-09-08
Views: 1249
|
1599552000 |
1249 |
 |
Description:
Author: hrbaylous
Session Name: Fall20 yeast test group 1
Genome Assembly: sacCer3
Creation Date: 2020-09-08
Views: 1585
|
1599552000 |
1585 |
 |
Description:
Author: Haley Yandrasitz
Session Name: Fall20 Mouse Test group#5
Genome Assembly: mm9
Creation Date: 2020-09-08
Views: 1376
|
1599552000 |
1376 |
 |
Description:
Author: jzhicks07
Session Name: CGEMS yeast JMH test
Genome Assembly: sacCer3
Creation Date: 2020-09-07
Views: 1455
|
1599465600 |
1455 |
 |
Description:
Author: biologyassignment
Session Name: Fall19 Yeast test (ACG)
Genome Assembly: sacCer3
Creation Date: 2020-09-07
Views: 1536
|
1599465600 |
1536 |
 |
Description:
Author: ArcherS
Session Name: Fall19 Yeast test A.S.
Genome Assembly: sacCer3
Creation Date: 2020-09-07
Views: 1418
|
1599465600 |
1418 |
 |
Description:
Author: federikr
Session Name: Fall20 TAS2R38 group#3
Genome Assembly: hg19
Creation Date: 2020-09-07
Views: 1307
|
1599465600 |
1307 |
 |
Description:
Author: perryts
Session Name: Fall20 Yeast Test (TSP)
Genome Assembly: sacCer3
Creation Date: 2020-09-07
Views: 1446
|
1599465600 |
1446 |
 |
Description:
Author: maddieevans9
Session Name: Fall20 Yeast test (ME)
Genome Assembly: sacCer3
Creation Date: 2020-09-07
Views: 1512
|
1599465600 |
1512 |
 |
Description:
Author: lauren_soleo
Session Name: TAS2R38 group 1
Genome Assembly: hg19
Creation Date: 2020-09-07
Views: 1470
|
1599465600 |
1470 |
 |
Description:
Author: brockse
Session Name: TAS2R38 group 1
Genome Assembly: hg19
Creation Date: 2020-09-07
Views: 1481
|
1599465600 |
1481 |
 |
Description:
Author: Chadtb220
Session Name: Fall19 Yeast test CTB
Genome Assembly: sacCer3
Creation Date: 2020-09-07
Views: 1520
|
1599465600 |
1520 |
 |
Description:
Author: AMOza
Session Name: LMM_Conservation_hg38
Genome Assembly: hg38
Creation Date: 2020-09-09
Views: 10291
|
1599638400 |
10291 |
 |
Description:
Author: kianayaghoobian
Session Name: Fall19 Yeast test KY
Genome Assembly: sacCer3
Creation Date: 2020-09-07
Views: 1685
|
1599465600 |
1685 |
 |
Description:
Author: KeelynCrossin
Session Name: Fall20 yeast test KGC
Genome Assembly: sacCer3
Creation Date: 2020-09-06
Views: 1414
|
1599379200 |
1414 |
 |
Description:
Author: JFormosa
Session Name: Hwk6Pt2
Genome Assembly: hg19
Creation Date: 2022-02-22
Views: 828
|
1645516800 |
828 |
 |
Description:
Author: embdukes
Session Name: Fall20 Yeast test (EDB)
Genome Assembly: sacCer3
Creation Date: 2020-09-06
Views: 1510
|
1599379200 |
1510 |
 |
Description:
Author: napace
Session Name: Fall19 Yeast test NAP
Genome Assembly: sacCer3
Creation Date: 2020-09-06
Views: 1622
|
1599379200 |
1622 |
 |
Description:
Author: federikr
Session Name: Fall20 Yeast Test KF
Genome Assembly: sacCer3
Creation Date: 2020-09-06
Views: 1488
|
1599379200 |
1488 |
 |
Description:
Author: JRozick147
Session Name: wuhCor1-1
Genome Assembly: wuhCor1
Creation Date: 2020-09-05
Views: 1904
|
1599292800 |
1904 |
 |
Description:
Author: lauren_soleo
Session Name: Fall20 Yeast test LS
Genome Assembly: sacCer3
Creation Date: 2020-09-04
Views: 1450
|
1599206400 |
1450 |
 |
Description: This session is of the new ALFA (Allele Frequency Aggregator) variants and allele frequency hub. Results from the Allele Frequency Aggregator (ALFA) project will help users interpret the biological impact of common and rare sequence variants. ALFA's initial release includes analysis of genotype data from ~100K unrestricted dbGaP subjects and provides high-quality allele frequency data now displayed on relevant dbSNP records. Eventually ALFA will analyze more than 1M subjects. The current version is for hg38/GRCh38 with work to include hg19/GRCh37 data. Read more about the ALFA project page here: https://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa
Author: brianlee
Session Name: ALFA_Trio
Genome Assembly: hg38
Creation Date: 2020-09-03
Views: 2014
|
1599120000 |
2014 |
 |
Description:
Author: brockse
Session Name: Fall20 Yeast Test SB
Genome Assembly: sacCer3
Creation Date: 2020-08-31
Views: 1614
|
1598860800 |
1614 |
 |
Description:
Author: Yuanyuan Zhang
Session Name: Mechanism of 5mC controlled Arabidopsis clock pace
Genome Assembly: hub_329263_araTha1
Creation Date: 2021-01-11
Views: 3666
|
1610352000 |
3666 |
 |
Description:
Author: mongerc
Session Name: mm10Suz12
Genome Assembly: mm10
Creation Date: 2020-08-28
Views: 1523
|
1598601600 |
1523 |
 |
Description: A stop codon (marked in red) found at the end of the METTL3 gene, indicating the end of translation, which proceeds right to left for this gene. Below, we can observe how conservation decreases dramatically after the Stop codon in the “PhyloP” data track.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: mettl3_stop
Genome Assembly: hg19
Creation Date: 2022-01-19
Views: 982
|
1642579200 |
982 |
 |
Description:
Author: alfordml
Session Name: Spring22 Yeast test MA
Genome Assembly: sacCer3
Creation Date: 2022-01-25
Views: 911
|
1643097600 |
911 |
 |
Description:
Author: gera912
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2020-08-11
Views: 1499
|
1597132800 |
1499 |
 |
Description:
Author: feng xue
Session Name: kdm4c
Genome Assembly: hg17
Creation Date: 2020-08-10
Views: 1800
|
1597046400 |
1800 |
 |
Description:
Author: shuynhs
Session Name: SH-61951083-MEDG580-UBC-2020
Genome Assembly: hg38
Creation Date: 2020-10-05
Views: 1492
|
1601884800 |
1492 |
 |
Description: Session of ASHG 2020 Figure 2. Displays Hi-C track format with custom display options (score normalization, display color, arc display).
Author: Lou
Session Name: ASHG2
Genome Assembly: hg19
Creation Date: 2020-10-05
Views: 1537
|
1601884800 |
1537 |
 |
Description: Session of ASHG 2020 Figure 1. Displays Hi-C track format with default settings. Additional tracks include UCSC Genes, ChIP-seq signal track from ENCODE, and GeneHancer clustered interactions.
Author: Lou
Session Name: ASHG1
Genome Assembly: hg19
Creation Date: 2020-10-05
Views: 1518
|
1601884800 |
1518 |
 |
Description:
Author: boothc
Session Name: Fish n Chips detailed
Genome Assembly: danRer10
Creation Date: 2020-08-04
Views: 1554
|
1596528000 |
1554 |
 |
Description:
Author: elencampoy
Session Name: HsInv0573
Genome Assembly: hg19
Creation Date: 2021-02-18
Views: 1230
|
1613635200 |
1230 |
 |
Description:
Author: jzhicks07
Session Name: CRX 9.28 JMH
Genome Assembly: hg38
Creation Date: 2020-09-28
Views: 1473
|
1601280000 |
1473 |
 |
Description:
Author: simonwang612
Session Name: Ying_ATAC_GSDMD
Genome Assembly: mm10
Creation Date: 2020-07-23
Views: 1929
|
1595491200 |
1929 |
 |
Description:
Author: simonwang612
Session Name: Ying_ATACseq_peaks
Genome Assembly: mm10
Creation Date: 2020-07-23
Views: 1953
|
1595491200 |
1953 |
 |
Description:
Author: simonwang612
Session Name: atac_Ying
Genome Assembly: mm10
Creation Date: 2020-07-23
Views: 1679
|
1595491200 |
1679 |
 |
Description:
Author: simonwang612
Session Name: mm10_atac_Ying
Genome Assembly: mm10
Creation Date: 2020-07-23
Views: 1502
|
1595491200 |
1502 |
 |
Description:
Author: Lachlan Coin
Session Name: dRNA
Genome Assembly: hg38
Creation Date: 2020-07-22
Views: 1572
|
1595404800 |
1572 |
 |
Description: Tracks associated with: Buzzi et al. 2022 Sox8 remodels the cranial ectoderm to generate the ear
Author: alexthiery
Session Name: Sox8_otic_reprogramming
Genome Assembly: galGal6
Creation Date: 2021-03-02
Views: 718
|
1614672000 |
718 |
 |
Description:
Author: Marie-José
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2020-07-28
Views: 1829
|
1595923200 |
1829 |
 |
Description:
Author: shaghayegh dastjerdi
Session Name: hg19/smad3 new promoters
Genome Assembly: hg19
Creation Date: 2020-07-27
Views: 1569
|
1595836800 |
1569 |
 |
Description:
Author: Schne2lk
Session Name: hg38 RHO multiomics
Genome Assembly: hg38
Creation Date: 2021-11-16
Views: 969
|
1637049600 |
969 |
 |
Description:
Author: ashleywilkins
Session Name: hg38 retinal multiomics
Genome Assembly: hg38
Creation Date: 2021-11-16
Views: 891
|
1637049600 |
891 |
 |
Description:
Author: chalie102
Session Name: HDA_ChIP_H3K18ac
Genome Assembly: sacCer3
Creation Date: 2021-02-15
Views: 1817
|
1613376000 |
1817 |
 |
Description:
Author: vperreau@unimelb.edu.au
Session Name: hg38_NTRK2_blat_probes
Genome Assembly: hg38
Creation Date: 2020-10-12
Views: 1715
|
1602489600 |
1715 |
 |
Description:
Author: LalehEbr
Session Name: hg19 UCSC workshop assignment
Genome Assembly: hg19
Creation Date: 2020-07-19
Views: 1938
|
1595145600 |
1938 |
 |
Description:
Author: fatemeh sadat
Session Name: F
Genome Assembly: hg19
Creation Date: 2020-07-19
Views: 1931
|
1595145600 |
1931 |
 |
Description:
Author: shaghayegh dastjerdi
Session Name: hg19/dastjerdi
Genome Assembly: hg19
Creation Date: 2020-07-17
Views: 2008
|
1594972800 |
2008 |
 |
Description:
Author: fatemeh sadat
Session Name: FASc
Genome Assembly: hg19
Creation Date: 2020-07-19
Views: 2167
|
1595145600 |
2167 |
 |
Description:
Author: shaghayegh dastjerdi
Session Name: hg19/Smad3 promoter
Genome Assembly: hg19
Creation Date: 2020-07-17
Views: 1591
|
1594972800 |
1591 |
 |
Description: Transcript structures for LncExpDB and LncBook databases
Author: lizhao
Session Name: lncexpdb_UCSC
Genome Assembly: hg38
Creation Date: 2023-09-07
Views: 161199
|
1694073600 |
161199 |
 |
Description: MetamORF: A repository of unique small Open Reading Frames identified by both experimental and computational approaches for gene-level and meta analyses
Author: schoteau
Session Name: MetamORF (hg38)
Genome Assembly: hg38
Creation Date: 2020-07-14
Views: 1570
|
1594713600 |
1570 |
 |
Description:
Author: mongerc
Session Name: Brien2020ChipRX
Genome Assembly: hg38
Creation Date: 2020-07-09
Views: 1548
|
1594281600 |
1548 |
 |
Description:
Author: mongerc
Session Name: Brien2020RNASeq
Genome Assembly: hg38
Creation Date: 2020-07-09
Views: 1626
|
1594281600 |
1626 |
 |
Description:
Author: mongerc
Session Name: Brien2020ATAC
Genome Assembly: hg38
Creation Date: 2020-07-09
Views: 1568
|
1594281600 |
1568 |
 |
Description:
Author: Marie-José
Session Name: hg19_TAD
Genome Assembly: hg19
Creation Date: 2020-07-09
Views: 1994
|
1594281600 |
1994 |
 |
Description:
Author: Stephanie
Session Name: hg19-OXTR
Genome Assembly: hg19
Creation Date: 2020-07-08
Views: 1577
|
1594195200 |
1577 |
 |
Description: RCRN
Author: Ruffi2ec
Session Name: Ruffing_ Lab 4-10 RCRN
Genome Assembly: mm9
Creation Date: 2024-04-10
Views: 289
|
1712736000 |
289 |
 |
Description:
Author: Stephanie
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2020-07-08
Views: 1580
|
1594195200 |
1580 |
 |
Description:
Author: Chen Chunpeng
Session Name: GBM_STAT
Genome Assembly: hg38
Creation Date: 2020-07-14
Views: 1505
|
1594713600 |
1505 |
 |
Description:
Author: Stephanie
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2020-07-08
Views: 1718
|
1594195200 |
1718 |
 |
Description:
Author: ranjan99
Session Name: RCas9_HSA_IM
Genome Assembly: mm10
Creation Date: 2020-06-28
Views: 1956
|
1593331200 |
1956 |
 |
Description:
Author: ustiugova_al
Session Name: cd27
Genome Assembly: hg38
Creation Date: 2020-06-26
Views: 1614
|
1593158400 |
1614 |
 |
Description:
Author: ccpowell
Session Name: solLy4
Genome Assembly: hub_2266061_solLy4
Creation Date: 2020-06-15
Views: 2445
|
1592208000 |
2445 |
 |
Description:
Author: Dbakalar
Session Name: Rapgef2 mRNA, promotors, H3K9me3
Genome Assembly: mm10
Creation Date: 2020-06-18
Views: 1765
|
1592467200 |
1765 |
 |
Description:
Author: Dbakalar
Session Name: Rapgef2 mRNA, promotors, H3K27me3
Genome Assembly: mm10
Creation Date: 2020-06-18
Views: 1536
|
1592467200 |
1536 |
 |
Description:
Author: Dbakalar
Session Name: Rapgef2 mRNA, promotors, H3K36me3
Genome Assembly: mm10
Creation Date: 2020-06-18
Views: 1504
|
1592467200 |
1504 |
 |
Description:
Author: Dbakalar
Session Name: Rapgef2 mRNA, promotors, H3K9ac
Genome Assembly: mm10
Creation Date: 2020-06-18
Views: 1450
|
1592467200 |
1450 |
 |
Description:
Author: Dbakalar
Session Name: Rapgef2 mRNA, promotors, H3K27ac
Genome Assembly: mm10
Creation Date: 2020-06-18
Views: 1588
|
1592467200 |
1588 |
 |
Description:
Author: Dbakalar
Session Name: Rapgef2 mRNA, promotors, K4me2
Genome Assembly: mm10
Creation Date: 2020-06-18
Views: 1479
|
1592467200 |
1479 |
 |
Description:
Author: Dbakalar
Session Name: Rapgef2 mRNA, promotors, H3Kme1
Genome Assembly: mm10
Creation Date: 2020-06-18
Views: 1429
|
1592467200 |
1429 |
 |
Description:
Author: Dbakalar
Session Name: Rapgef2 mRNA, promotors, H4me3
Genome Assembly: mm10
Creation Date: 2020-06-18
Views: 1516
|
1592467200 |
1516 |
 |
Description:
Author: jeramiahsmith
Session Name: RNAseq_regen_compil
Genome Assembly: hub_1106737_Amex_PQ.v4
Creation Date: 2020-06-17
Views: 1829
|
1592380800 |
1829 |
 |
Description:
Author: williamhconrad
Session Name: hg19 HCT116 DMR Methylation
Genome Assembly: hg19
Creation Date: 2020-06-09
Views: 1782
|
1591689600 |
1782 |
 |
Description:
Author: nickeener
Session Name: SARS-CoV-2 Selection
Genome Assembly: wuhCor1
Creation Date: 2020-06-03
Views: 1572
|
1591171200 |
1572 |
 |
Description:
Author: br1215@nyu.edu
Session Name: ce10_OP_CNV
Genome Assembly: ce10
Creation Date: 2020-06-01
Views: 1703
|
1590998400 |
1703 |
Screenshot not available
Click Here to view |
Description:
Author: dianegirard
Session Name: H3K27ac_ChIPseq_EPI_hg38
Genome Assembly: hg38
Creation Date: 2020-06-01
Views: 1818
|
1590998400 |
1818 |
Screenshot not available
Click Here to view |
Description:
Author: dianegirard
Session Name: H3K27ac_ChIPseq_EPICARD_hg38
Genome Assembly: hg38
Creation Date: 2020-05-30
Views: 1919
|
1590825600 |
1919 |
Screenshot not available
Click Here to view |
Description:
Author: dianegirard
Session Name: H3K27ac_ChIPseq_CARD
Genome Assembly: hg38
Creation Date: 2020-05-30
Views: 1607
|
1590825600 |
1607 |
 |
Description: Zoomed in on the first exon of the METTL3 gene. If we look at the direction of translation/arrows on the introns, we can see that translation for this gene is going from right to left. Zooming in here, the methionine start codon, colored in green, is observable. Notice how the sequence upstream from the start codon
(to the right) is an untranslated region, depicted as a half-height box at the UTR region of the 5’ end. Offscreen to the right is more non-coding RNA (indicated by double white arrows). Only after the start codon does translation actually begin in the right-to-left direction.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: mettl3_exon1
Genome Assembly: hg19
Creation Date: 2022-01-19
Views: 1008
|
1642579200 |
1008 |
 |
Description: ChIPmera
Author: CGRoque
Session Name: GouveiaRoque_ChIPmera
Genome Assembly: hg38
Creation Date: 2021-04-22
Views: 788
|
1619078400 |
788 |
 |
Description:
Author: ccpowell
Session Name: solLyc1
Genome Assembly: hub_2224691_solLyc1
Creation Date: 2020-05-25
Views: 1710
|
1590393600 |
1710 |
 |
Description:
Author: br1215@nyu.edu
Session Name: ce10_CNV_10kb_FINAL
Genome Assembly: ce10
Creation Date: 2020-05-17
Views: 1686
|
1589702400 |
1686 |
 |
Description:
Author: br1215@nyu.edu
Session Name: ce10_CNV_FINAL
Genome Assembly: ce10
Creation Date: 2020-05-17
Views: 1638
|
1589702400 |
1638 |
 |
Description:
Author: m1kss
Session Name: mouse chr16 methylation embryonic
Genome Assembly: mm10
Creation Date: 2020-05-13
Views: 1655
|
1589356800 |
1655 |
 |
Description:
Author: yael porat
Session Name: myc
Genome Assembly: hg38
Creation Date: 2020-05-10
Views: 1614
|
1589097600 |
1614 |
 |
Description: RNA plus-sense secondary structure prediction by Rangan et al are displayed. Both MEA secondary structured and MSA conserved sequences are formatted as separate tracks.
All instances of "CUAAAC" motifs are annotated (plus-sense purple, negative-sense teal, complement GUUUAG)
Yellow highlights represent hypothesized negative or complement interactions. Orange highlights represent non-canonical template switching overlapping with regions with high confidence secondary structure predictions. Blue highlights represent canonical TRS-L TRS-B template switching.
Author: jssim
Session Name: wuhCor
Genome Assembly: wuhCor1
Creation Date: 2020-04-23
Views: 1764
|
1587628800 |
1764 |
 |
Description:
Author: apacis
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2020-05-06
Views: 2168
|
1588752000 |
2168 |
 |
Description:
Author: Annaneed
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2020-07-06
Views: 1559
|
1594022400 |
1559 |
 |
Description:
Author: tsvid
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2021-09-27
Views: 1087
|
1632729600 |
1087 |
 |
Description:
Author: aoludipe
Session Name: hg19_kishimoto 1998
Genome Assembly: hg19
Creation Date: 2021-09-27
Views: 1400
|
1632729600 |
1400 |
 |
Description: All patient isolates in PRJNA613958, with exception of MinION sequenced, were individually aligned against the NC_045512.2 reference with SNPs called for each isolate using 'ngskit4b kalign'. A matrix of all individual isolate SNP site loci (rows) and isolate base calls at that loci (cols) created with 'ngskit4b snpmarkers' and this matrix then processed with 'ngskit4b snps2pgsnps' to output the pgSNPs tracks. Tracks show, for each SNP loci, the number of isolates containing the displayed allele base. Difference in tracks results from strictness of parameterisations (Allelic PValue, read coverage) used when calling SNPs.
Author: biodiscovery
Session Name: wuhCor1 PRJNA613958 allele union
Genome Assembly: wuhCor1
Creation Date: 2020-04-26
Views: 1774
|
1587888000 |
1774 |
 |
Description:
Author: tmitchell1
Session Name: SNCG with EXON2 CT and CHB and SK-N-MC
Genome Assembly: hg38
Creation Date: 2020-04-15
Views: 1580
|
1586937600 |
1580 |
 |
Description:
Author: tmitchell1
Session Name: SNCB CT with CHB and SK-N-MC
Genome Assembly: hg38
Creation Date: 2020-04-15
Views: 1989
|
1586937600 |
1989 |
 |
Description:
Author: tmitchell1
Session Name: SNCG CT with CHB and SK-N-MC
Genome Assembly: hg38
Creation Date: 2020-04-15
Views: 1711
|
1586937600 |
1711 |
 |
Description:
Author: ssdhar
Session Name: hg19 new H3k27 ac Her3
Genome Assembly: hg19
Creation Date: 2021-08-16
Views: 1263
|
1629100800 |
1263 |
 |
Description:
Author: kianayaghoobian
Session Name: CRISPR
Genome Assembly: hg38
Creation Date: 2020-09-28
Views: 1454
|
1601280000 |
1454 |
 |
Description:
Author: tmitchell
Session Name: WT and mutant on CFTR
Genome Assembly: mm9
Creation Date: 2020-04-03
Views: 1717
|
1585900800 |
1717 |
 |
Description:
Author: tmitchell
Session Name: WT and Mutant on Guca1b
Genome Assembly: mm9
Creation Date: 2020-04-03
Views: 1535
|
1585900800 |
1535 |
 |
Description:
Author: tmitchell
Session Name: WT and mutant on Guca1a
Genome Assembly: mm9
Creation Date: 2020-04-03
Views: 1727
|
1585900800 |
1727 |
 |
Description:
Author: tmitchell
Session Name: WT and Mutant CRX
Genome Assembly: mm9
Creation Date: 2020-04-03
Views: 1584
|
1585900800 |
1584 |
 |
Description:
Author: tmitchell
Session Name: MAPT mutation custom track
Genome Assembly: hg38
Creation Date: 2020-04-03
Views: 1606
|
1585900800 |
1606 |
 |
Description:
Author: tmitchell
Session Name: Exonic seq Custom Track
Genome Assembly: hg19
Creation Date: 2020-04-03
Views: 1623
|
1585900800 |
1623 |
 |
Description:
Author: liaohy
Session Name: RNAseq_Xia.L_Control_Cex
Genome Assembly: hg19
Creation Date: 2020-04-01
Views: 1690
|
1585728000 |
1690 |
 |
Description:
Author: liaohy
Session Name: RNAseq_Lin.XT_AM20191204
Genome Assembly: hg38
Creation Date: 2020-04-01
Views: 1741
|
1585728000 |
1741 |
 |
Description:
Author: Genomedx
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2020-04-09
Views: 2062
|
1586419200 |
2062 |
 |
Description:
Author: tmitchell
Session Name: WT and Mutant Arr3
Genome Assembly: mm9
Creation Date: 2020-04-03
Views: 1574
|
1585900800 |
1574 |
 |
Description:
Author: andre2tj
Session Name: 3/31_Lab_Assignment_1
Genome Assembly: hg19
Creation Date: 2020-03-31
Views: 1818
|
1585641600 |
1818 |
 |
Description:
Author: liaohy
Session Name: RNAseq_Lian.JB_AM20200311-01
Genome Assembly: hg38
Creation Date: 2020-03-30
Views: 1682
|
1585555200 |
1682 |
 |
Description:
Author: sajjeev
Session Name: ICR_CMYC_optimization_03262020
Genome Assembly: hg19
Creation Date: 2020-03-26
Views: 1845
|
1585209600 |
1845 |
 |
Description: Hippo-YAP signaling controls lineage differentiation of mouse embryonic stem cells through modulating the formation of super-enhancers
Author: ZJRen
Session Name: Yap_RNAseq_part1
Genome Assembly: mm10
Creation Date: 2020-03-25
Views: 817
|
1585123200 |
817 |
 |
Description: Hippo-YAP signaling controls lineage differentiation of mouse embryonic stem cells through modulating the formation of super-enhancers
Author: ZJRen
Session Name: Yap_ChIPseq_part2
Genome Assembly: mm10
Creation Date: 2020-03-25
Views: 826
|
1585123200 |
826 |
 |
Description: Hippo-YAP signaling controls lineage differentiation of mouse embryonic stem cells through modulating the formation of super-enhancers
Author: ZJRen
Session Name: Yap_ChIPseq_part1
Genome Assembly: mm10
Creation Date: 2020-03-25
Views: 858
|
1585123200 |
858 |
 |
Description:
Author: ChPreuss
Session Name: MODEL_AD_NONCODING_VARIANTS_PRIORITIZATION
Genome Assembly: hg19
Creation Date: 2020-03-24
Views: 1773
|
1585036800 |
1773 |
 |
Description: ReMap 2020: An atlas of regulatory regions from an integrative analysis of Human and Arabidopsis thaliana DNA-binding sequencing experiments. Go to <a href="http://remap.univ-amu.fr">ReMap</a> for more info.
Author: Benoit Ballester
Session Name: US_ReMap2020_hg38
Genome Assembly: hg38
Creation Date: 2019-08-28
Views: 4558
|
1566979200 |
4558 |
 |
Description: METTL3 gene on human assembly hg38, showing a gene transcribed from the negative, or lower strand. You can see the exon numbering if you mouse over the exons on either end of the gene. The 5-end of the gene is on the right.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg38_revStrandGene
Genome Assembly: hg38
Creation Date: 2022-05-31
Views: 954
|
1653984000 |
954 |
 |
Description:
Author: fengtao-cau
Session Name: susScr11
Genome Assembly: susScr11
Creation Date: 2020-03-10
Views: 1798
|
1583827200 |
1798 |
 |
Description:
Author: apodgorny
Session Name: RNA_Seq
Genome Assembly: hg38
Creation Date: 2021-08-04
Views: 1211
|
1628064000 |
1211 |
 |
Description: “Spliced ESTs” data track set to pack showing varying gene arrangements for the FGFR2 gene. Each EST is labeled with the corresponding tag to the left of its sequence. CN402795 sequence read is missing exon 6 for the FGFR2 gene (yellow highlight). In the orange highlighted region, we can see two more splice variants BE706533 and BE147547 missing exon 7.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: fgfr2_highlights
Genome Assembly: hg19
Creation Date: 2022-02-28
Views: 902
|
1646035200 |
902 |
 |
Description:
Author: jeramiahsmith
Session Name: JS_structure_02262020
Genome Assembly: hub_1106737_Amex_PQ.v4
Creation Date: 2020-02-26
Views: 1543
|
1582704000 |
1543 |
 |
Description:
Author: celiagonzalez98
Session Name: Team Work 2
Genome Assembly: hg19
Creation Date: 2020-02-24
Views: 1758
|
1582531200 |
1758 |
 |
Description:
Author: celiagonzalez98
Session Name: Team Work
Genome Assembly: hg38
Creation Date: 2020-02-24
Views: 1638
|
1582531200 |
1638 |
 |
Description:
Author: Fu Haifeng
Session Name: Primed VS Naive
Genome Assembly: mm10
Creation Date: 2020-02-21
Views: 1633
|
1582272000 |
1633 |
 |
Description:
Author: chend8
Session Name: ATAC_150_intergenic_5
Genome Assembly: dm6
Creation Date: 2020-02-19
Views: 2020
|
1582099200 |
2020 |
 |
Description:
Author: vinayswamy
Session Name: DNTX
Genome Assembly: hg38
Creation Date: 2020-02-17
Views: 1779
|
1581926400 |
1779 |
 |
Description:
Author: Tamira
Session Name: Mouse ECR
Genome Assembly: hg19
Creation Date: 2020-02-17
Views: 1828
|
1581926400 |
1828 |
 |
Description:
Author: gaguil17
Session Name: Intersection.
Genome Assembly: mm10
Creation Date: 2020-02-16
Views: 1640
|
1581840000 |
1640 |
 |
Description:
Author: celiagonzalez98
Session Name: class 4 part 2
Genome Assembly: hg19
Creation Date: 2020-02-13
Views: 1903
|
1581580800 |
1903 |
 |
Description:
Author: celiagonzalez98
Session Name: class 4 part 1
Genome Assembly: hg19
Creation Date: 2020-02-13
Views: 1919
|
1581580800 |
1919 |
 |
Description:
Author: UxueHuarte
Session Name: my sesion
Genome Assembly: hg19
Creation Date: 2020-02-13
Views: 1614
|
1581580800 |
1614 |
 |
Description:
Author: dmr2928
Session Name: Daniel Robertson
Genome Assembly: mm10
Creation Date: 2020-02-12
Views: 1677
|
1581494400 |
1677 |
 |
Description:
Author: AdrianFer
Session Name: Intersected_Updated
Genome Assembly: mm10
Creation Date: 2020-02-12
Views: 1741
|
1581494400 |
1741 |
 |
Description:
Author: gaguil17
Session Name: Fgf17
Genome Assembly: mm10
Creation Date: 2020-02-12
Views: 2015
|
1581494400 |
2015 |
 |
Description:
Author: ManteoAndrade
Session Name: bejeranofoxp2
Genome Assembly: hg19
Creation Date: 2020-02-12
Views: 1652
|
1581494400 |
1652 |
 |
Description:
Author: celiagonzalez98
Session Name: class 3 part 2
Genome Assembly: hg19
Creation Date: 2020-02-12
Views: 1630
|
1581494400 |
1630 |
 |
Description:
Author: celiagonzalez98
Session Name: class 2 enviar
Genome Assembly: hg38
Creation Date: 2020-02-11
Views: 1906
|
1581408000 |
1906 |
 |
Description:
Author: mecay.1
Session Name: Session 2 final
Genome Assembly: hg38
Creation Date: 2020-02-11
Views: 1709
|
1581408000 |
1709 |
 |
Description:
Author: JonMatas
Session Name: GenoPractice 2 Public
Genome Assembly: hg38
Creation Date: 2020-02-11
Views: 1878
|
1581408000 |
1878 |
 |
Description: Juniata Bioinformatics
Infantile Parkinsonism – dystonia is the second most popular neurodegenerative disorder. It is followed by Alzheimer. This disease is present in infancy and prevent those affected to live past their teenage year. This disease is fatal and has a second name known as Transporter deficiency Syndrome (DTDS). Those individuals that are affected may have many problem; limb dyskinesia, dystonia, and chorea, (OMIM,1). Currently we do not have an effective treatment to treat Infantile Parkinsonism-dystonia. This genetic mutation shows an increase between HVA and 5-HIAA in the cerebrospinal fluid. This leads to an increase in dopamine to serotonin metabolites, (OMIN,1).
Author: mmendez
Session Name: Juniata Bioinformatics; MM & NP
Genome Assembly: hg19
Creation Date: 2020-02-10
Views: 1344
|
1581321600 |
1344 |
 |
Description: Set of publicly available tracks to define enhance regions used in the human left ventricle. Accompanies Gacita et al, 2020.
Author: agacita
Session Name: Gacita_et_al_LV_Enhancer_Tracks
Genome Assembly: hg19
Creation Date: 2020-02-10
Views: 2398
|
1581321600 |
2398 |
 |
Description:
Author: Andy20UNAV
Session Name: SESION2
Genome Assembly: hg38
Creation Date: 2020-02-11
Views: 1969
|
1581408000 |
1969 |
 |
Description:
Author: irish
Session Name: Example_Diana_gtfassemblyW1_w2
Genome Assembly: hg38
Creation Date: 2020-02-10
Views: 2570
|
1581321600 |
2570 |
 |
Description: Juniata Bioinformatics BMKJ The original encoded protein functions as a transcriptional coactivator that increases c-Myc activity and inhibits transforming growth factor-beta (TGF-beta) and nuclear factor kappa-B (NF-kB) signaling. The encoded protein also regulates the stability of cyclin D1 mRNA and may play a role in cell proliferation and cancer progression. There is only one novel variant of this gene, and it is GLU366GLY. In the Puffenberger paper, they used a tool called PolyPhen-2, which can be found on Harvard's website, said the substitution was "probably damaging" with a score of 0.99. Some common characteristics expressed from the mutation are psychomotor delay, intractable seizures/epilepsy, bulbous nose, wide mouth and tongue, broad jaw, short hands, short tapered fingers, and broad thumbs.
The first track pictured below shows a graph that serves as a basis for the predicted alternative splicing transcripts shown in the SIB Genes track. The blocks represent exons and the lines indicate introns. Also, there is a vertex for each splice, start, and end.
The graph under the previous track shows the relationship between genetic variation and gene expression in multiple human tissues.
Author: bmiller913
Session Name: SNIP1 BMKJ
Genome Assembly: hg19
Creation Date: 2020-02-09
Views: 1668
|
1581235200 |
1668 |
 |
Description: Children with Usher’s syndrome show no problems during infancy but begin to lose their ability to hear and see during childhood. Patients also present with mild lower body abnormalities. Charles Bonnet syndrome may also occur after affected children contract an infectious illness. Charles Bonnet syndrome is characterized by “decreased visual acuity and vivid visual hallucinations.” Usher’s syndrome is caused by a missense mutation on the 454 position of the Histidyl-tRNA synthetase (HARS) gene in which a tyrosine is replaced with an serine. The normal function of HARS is to charge tRNA with a histidine amino acid. Evidence suggests that the mutation that causes Usher’s syndrome may change the recognition of tRNA codons and/or catalytic activity.
The Conservation track shows that this region is conserved across several species, and has amino acid variation in two of these species: Zebrafish (M instead of Y) and Lamprey (V instead of Y). Conservation across a number of species suggests that this region is essential to many vertebrates, and is perhaps why a mutation of this region is so detrimental.
The CRISPR targets track shows DNA sequences that are targetable by CRISPR RNA guides. This track is able to show an estimate of how efficient the cleavage would be at the site (Green is the highest) as well as how specific the 20-mer primer is to a specific site in the genome (Black is least specific to just that site). This could be helpful when exploring options for gene therapy or modifications to the gene.
The ClinVar Variants track shows the positions of variants in the ClinVar database. This track comes with two subtracks: one for long variants (100 bp or more, most of which are copy number variants) and one for short variants (less than 100 bp). The short variant track color codes variants according to their pathogenicity. Pathogenic variants are colored red, benign variants are colored green, and variants of uncertain significance are colored gray. The long variant track color code variants according to whether the variant is a copy number gain or loss. A user can click on a variant to find more information about the variant including a report that contains an interpretation of the variant and peer reviewed citations.
Author: behreka20
Session Name: Juniata Bioinformatics KB, LL, CM
Genome Assembly: hg19
Creation Date: 2020-02-09
Views: 1684
|
1581235200 |
1684 |
 |
Description: The tracks in the Synonymous Constraint Public Hub represent regions of protein-coding sequences in which the rate of synonymous substitutions during mammalian evolution has been significantly lower or higher than in the rest of that gene. There are two tracks: Synonymous Constraint Elements (SCEs) have had lower than normal synonymous substitution rates while Synonymous Acceleration Elements (SAEs) have had higher than normal rates. Thanks to Maxim Wolf, Irwin Jungreis and others at MIT for the hub.
Author: PublicSessions
Session Name: SynonymousConstraintHub
Genome Assembly: hg19
Creation Date: 2020-02-07
Views: 1942
|
1581062400 |
1942 |
 |
Description: Various mutations of the gene TUBGCP6 lead to alterations in the amino acid sequence of the subsequent protein, which leads to potential loss-of-function of the protein. This presents pathologically with ubiquitous marked microcephaly, a receding forehead, diminutive anterior fontanelle, and sutural ridging, in addition to other varied symptoms such as cognitive impairment, reduced cranial circumference, and epilepsy.
Author: cadeemlet
Session Name: Juniata Bioinformatics CE & SA
Genome Assembly: hg19
Creation Date: 2020-02-07
Views: 1416
|
1581062400 |
1416 |
 |
Description: Juniata Bioinformatics SA and CE. Various mutations in gene TUBGCP6 result in microcephaly and chorioretinopathy. This gene codes for a protein that is part of the gamma tubulin complex, which is required for microtubule nucleation at the centrosome, and thus mitosis beginning with interphase. Mutations affecting this protein can lead to symptoms of a receding forehead, diminutive anterior fontanelle, sutural ridging, cognitive delay, visual impairment, diffuse pachygyria, and a hypoplastic cerebellar vermis.
SA track: ENCODE Proteogenomics- this track is used to show functional elements of the human genome by using mass spectrometry to determine if translation is occurring by measuring protein produced by transcripts via proteogenomic mapping. This track identifies one exon within the TUBGCP6 gene. It is near the end of the mapped gene and its name is LGAGQTPGELLNPLVLNSVLSK.
CE track: GTEx RNA-seq- Displayed also is track for GTEx Gene, which displays the relative gene expression through total mRNA counts of TUBGCP6 in 53 tissue sites distributed throughout the body. What can be derived primarily through this track is that the tissues which exhibit the greatest effects of TUBGCP6 gene mutations also exhibit the highest levels of genetic expression of the gene in healthy individuals, indicating a more prominent role in affected regions.
Author: slca1991
Session Name: Juniata Bioinformatics SA and CE
Genome Assembly: hg19
Creation Date: 2020-02-07
Views: 1652
|
1581062400 |
1652 |
 |
Description:
Author: prfreire
Session Name: MOLM13 MEIS1 pull down
Genome Assembly: hg19
Creation Date: 2019-09-11
Views: 1117
|
1568188800 |
1117 |
 |
Description:
Author: ignaciogoyache
Session Name: dia5
Genome Assembly: hg38
Creation Date: 2020-02-14
Views: 1933
|
1581667200 |
1933 |
 |
Description:
Author: Joseph
Session Name: ICH_rat
Genome Assembly: hg38
Creation Date: 2020-02-03
Views: 1637
|
1580716800 |
1637 |
 |
Description:
Author: philip.tatman
Session Name: sacCer3RepTimingForReview
Genome Assembly: sacCer3
Creation Date: 2020-01-31
Views: 1670
|
1580457600 |
1670 |
 |
Description:
Author: philip.tatman
Session Name: sacCer3CDKByPassForReview
Genome Assembly: sacCer3
Creation Date: 2020-01-31
Views: 1620
|
1580457600 |
1620 |
 |
Description:
Author: dramirez
Session Name: NomLeu3PROseq1st
Genome Assembly: nomLeu3
Creation Date: 2020-04-22
Views: 1753
|
1587542400 |
1753 |
 |
Description: The nucleosome sequencing data of mouse brain NAC
cells from NCBI GEO archive accession GSE54263. The sequence alignment (mm9) coverage profiles (bigWig) of control, stress-susceptible and stress-resilient mice and the nucleosome occupancy change and shift events (BED) are shown.
Author: Erinija
Session Name: mouse NAC nucleosome events (mm9)
Genome Assembly: mm9
Creation Date: 2020-01-27
Views: 2035
|
1580112000 |
2035 |
 |
Description:
Author: philip.tatman
Session Name: hg38NocReleaseForReview
Genome Assembly: hg38
Creation Date: 2020-01-31
Views: 1605
|
1580457600 |
1605 |
 |
Description: The disease of interest is Infantile Parkinsonism found on the SLC6A3 gene as the variant IVS9+1G>T. Infantile Parkinsonism is described as is an extremely rare inherited neurological syndrome that presents in early infancy with hypo-kinetic Parkinsonism and dystonia that can lead to fatality. Parkinsonism is a condition that causes movement abnormalities seen in Parkinson's disease such as tremors, slow movement, and impaired speech or muscle stiffness due to he loss of dopamine-containing neurons. Dystonia is a movement disorder in which a person's muscles contract uncontrollably. The SCL6A3 gene provides instruction for making proteins, specifically a dopamine transporter. This protein is found in neurons and is responsible for the transport of dopamine into the cells. Dopamine is a chemical messenger that signals for motivation, behavior, and control movement. The variant changes the first nucleotide of the ninth intron from a G to a T. This mutation would result in a change of the splicing pattern of the DNA.
The track that Nicci explored was the CRISPR Targets track which shows the DNA sequences targetable by CRISPR RNA guides using the enzyme Cas9 over the whole human genome. CRISPR technology is a simple yet powerful tool for editing genomes. It allows researchers to easily alter DNA sequences and modify gene function. The track shows all potential -NGG target sites across the genome. Gray or black regions indicate impossible or hard to target areas. The colors blue, red, yellow, and green indicate the predicted chance of cleavage from unable to calculate to high predicted cleavage respectively. Mel explored the Vega Genes track which shows gene annotations from the Vertebrate Genome Annotation (Vega) database. Annotations are divided into two sub tracks from the Vega Human Genome Annotation project. The Two sub tracks include Vega protein coding and noncoding gene annotations as well as Vega annotated pseudogenes and immunoglobulin segments. This track follows the display conventions for gene prediction tracks. Transcript type may be found by clicking on the transcript identifier which forms the outside link to the Vega link detail page.
School: Juniata College
Author: NMP94876
Session Name: Homewrok 3 Nicole Piccioni and Mell ended Juanita College Bioinformatics
Genome Assembly: hg19
Creation Date: 2020-02-29
Views: 1576
|
1582963200 |
1576 |
 |
Description:
Author: keconne
Session Name: TAS2R38 group#5
Genome Assembly: hg19
Creation Date: 2020-01-21
Views: 1453
|
1579593600 |
1453 |
 |
Description:
Author: nguye5vx
Session Name: Spring 2020 TAS Test VN
Genome Assembly: hg19
Creation Date: 2020-01-21
Views: 1551
|
1579593600 |
1551 |
 |
Description:
Author: dschmelt
Session Name: Han_Chinese_LCT_Gene
Genome Assembly: hub_1969921_GCA_002180035.3_HG00514_prelim_3.0
Creation Date: 2020-01-20
Views: 1887
|
1579507200 |
1887 |
 |
Description:
Author: keough
Session Name: monkey_guides
Genome Assembly: chlSab2
Creation Date: 2020-06-30
Views: 1187
|
1593504000 |
1187 |
 |
Description:
Author: Maksimov_Denis
Session Name: ChromHMM_clusters
Genome Assembly: hg19
Creation Date: 2021-05-11
Views: 1102
|
1620720000 |
1102 |
 |
Description:
Author: swttalyan
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2020-02-20
Views: 1634
|
1582185600 |
1634 |
 |
Description:
Author: ilasa.5
Session Name: Day 3 group 5
Genome Assembly: hg19
Creation Date: 2020-02-12
Views: 1703
|
1581494400 |
1703 |
 |
Description:
Author: ManteoAndrade
Session Name: foxp2
Genome Assembly: hg19
Creation Date: 2020-02-12
Views: 1606
|
1581494400 |
1606 |
 |
Description:
Author: keconne
Session Name: CGEMS yeast KEC test
Genome Assembly: sacCer3
Creation Date: 2020-01-15
Views: 1626
|
1579075200 |
1626 |
 |
Description:
Author: tmitchell
Session Name: Spring 2020 yeast test TM
Genome Assembly: sacCer3
Creation Date: 2020-01-15
Views: 1584
|
1579075200 |
1584 |
 |
Description:
Author: marti4sm
Session Name: Fall19 Yeast test SM
Genome Assembly: sacCer3
Creation Date: 2020-01-15
Views: 1990
|
1579075200 |
1990 |
 |
Description: This was an example of a CyVerse hosted assembly hub at the PAG 2020 workshop. The data at CyVerse is hosted at this directory that you can click and see the files: <a href="https://data.cyverse.org/dav-anon/iplant/home/brianleesoe/AssemblyHub_Ex1/">https://data.cyverse.org/dav-anon/iplant/home/brianleesoe/AssemblyHub_Ex1/</a>. In that directory you can click and read the text file named <a href="https://data.cyverse.org/dav-anon/iplant/home/brianleesoe/AssemblyHub_Ex1/hub.txt">hub.txt</a> and see how an assembly hub is a few stanzas that define the names of a genome and where to find binary-indexed raw data to display on the browser. The data is defined by the <code>twoBitPath araTha1.2bit</code> line in the genome stanza and in the <code>bigDataUrl cytoBandIdeo.bigBed</code> and <code>bigDataUrl ensGene.araTha1.bb</code> lines in the track stanzas. There is also an index file on the genes track <code>searchTrix ensGene.araTha1.ix</code> that allows to search the genes by both their names, such as <b> abcc2 </b>and their accessions, such as <b>AT3G50470.1</b>.
Author: brianlee
Session Name: CyVerse_Assembly_Hub
Genome Assembly: hub_1979095_araTha1
Creation Date: 2020-01-13
Views: 1611
|
1578902400 |
1611 |
 |
Description: This session reflects a PAG 2020 Sunday Session, Genomics of Non-Classical Model Animals, and a speaker Bridgett vonHoldt from Princeton University. The title of Dr. vonHoldt's talk was <b>Everyone Is Your Friend! the Molecular Architecture of Hypersocial Canines</b>. Dr. vonHoldt analyzed wolf and dog populations and discovered a 5-Mb genomic region on chromosome 6 in dogs previously found to be under positive selection in domestic dog breeding. They gathered data on friendliness between a captive wolf and pet dog populations and then examined variants. The region affected by structural variants associated with the exuberant sociability of domestic dogs related to a gene WBSCR17 on chr7 in humans linked to Williams-Beuren syndrome (WBS), a multisystem congenital disorder characterized by hypersocial behavior. Humans with this mutation have a lack of stranger danger and are overly cheerful. This session shows the WBSCR17 gene in human.
Author: brianlee
Session Name: Human_WBSCR17
Genome Assembly: hg38
Creation Date: 2020-01-13
Views: 1676
|
1578902400 |
1676 |
 |
Description: This session reflects a PAG 2020 Sunday Session, Genomics of Non-Classical Model Animals, and a speaker Claudio V. Mello from Oregon Health and Science University. Dr. Mello's talk was titled <b>What Comparative Studies of Parrot Genomes Can Teach Us about Longevity, Large Brains and Cognition</b>. Dr. Mello identified genomic features under selection in parrots and other long-lived birds that included telomerase activity (TERT), DNA damage repair, control of cell proliferation, cancer, immunity, and anti-oxidative mechanisms. His group also identified many brain-expressed, parrot-specific paralogs with known functions in neural development or vocal-learning brain circuits seen in humans (AUS2). This session shows similar alignment information in the conservation track around human AUTS2 as in one of Dr. Mello’s supplemental slides.
Author: brianlee
Session Name: Human_AUTS2
Genome Assembly: hg38
Creation Date: 2020-01-13
Views: 1863
|
1578902400 |
1863 |
 |
Description: This session reflects a PAG 2020 Poster by Hao Meng et al. from Peking-Tsinghua Center for Life Sciences, Peking University. The title of the poster was <b>Morphology and Genetics of Kinked Tails of Domestic Cats (Felis catus) in East and Southeast Asia</b>. Hao Meng and colleagues investigated the morphology and genetics of kinked tails in cats. Mutations in coding sequences (CDS) of T and HES7 were identified correlated to the short tails in Manx and Japanese bobtail breeds as well as some feral cats in Asia, However, about one third of short-tailed cats in China do not carry either variant. The poster described a new 1.6 Mb region with non-synonymous mutations were seen, suggesting changes in regulatory regions, the poster concluded.
An example of viewing this region on the UCSC Browser with the regulatory Gene Interactions track turned on. In humans the interactions track seem to suggest this region has many long distant interactions spanning across it from distant enhancers.
Author: brianlee
Session Name: Human_HES7
Genome Assembly: hg38
Creation Date: 2020-01-13
Views: 1688
|
1578902400 |
1688 |
 |
Description: This session reflects a PAG 2020 Saturday Session, Cattle/Sheep/Goat 1, and the speaker George E. Liu from the Animal Genomics and Improvement Laboratory, USDA-ARS. The session title was <b>Comparative Epigenomics and Genotype-Phenotype Association Analyses Revealed Conserved Genetic Architecture underlying Complex Traits between Cattle and Human</b>. Dr. Liu’s lab examined comparative genomics around the histone marks and found that they could transfer cell-type-specific information from the human data to infer similar issues about genes related to certain diseases. A specific example was PIK3CG, a gene high specifically expressed in mononuclear cells was significantly associated with both age-at-menopause in human and daugter-still-birth in cattle. This session shows PIK3CG in human.
Author: brianlee
Session Name: Human_PIK3CG
Genome Assembly: hg38
Creation Date: 2020-01-13
Views: 1735
|
1578902400 |
1735 |
 |
Description:
Author: ManteoAndrade
Session Name: mi ventana
Genome Assembly: hg38
Creation Date: 2020-02-11
Views: 2011
|
1581408000 |
2011 |
 |
Description:
Author: nguye5vx
Session Name: Fall19 Yeast Test VN
Genome Assembly: sacCer3
Creation Date: 2020-01-19
Views: 1747
|
1579420800 |
1747 |
 |
Description:
Author: agacita
Session Name: enhancer_dominic
Genome Assembly: hg19
Creation Date: 2020-01-03
Views: 1685
|
1578038400 |
1685 |
 |
Description:
Author: saadiak2
Session Name: MS
Genome Assembly: mm9
Creation Date: 2019-12-19
Views: 1719
|
1576742400 |
1719 |
 |
Description:
Author: celiagonzalez98
Session Name: class 2
Genome Assembly: hg38
Creation Date: 2020-02-11
Views: 1950
|
1581408000 |
1950 |
 |
Description:
Author: mecay.1
Session Name: session 2
Genome Assembly: hg38
Creation Date: 2020-02-11
Views: 1896
|
1581408000 |
1896 |
 |
Description: ATAC-seq of exocrine epithelial stem cells
Author: woaichixiangcai
Session Name: Exocrine_ATAC
Genome Assembly: mm10
Creation Date: 2024-06-29
Views: 305
|
1719648000 |
305 |
 |
Description:
Author: katerynamarku
Session Name: TAS2R38
Genome Assembly: hg19
Creation Date: 2022-02-01
Views: 817
|
1643702400 |
817 |
 |
Description:
Author: JonMatas
Session Name: Geno3 G4
Genome Assembly: hg19
Creation Date: 2020-02-12
Views: 1984
|
1581494400 |
1984 |
 |
Description:
Author: egrossi
Session Name: Hi-C BAF47
Genome Assembly: hg18
Creation Date: 2019-11-29
Views: 1888
|
1575014400 |
1888 |
 |
Description: In this specific session of 3 bases with scores missing, at the very end of chrM of mm10, you can see data is lacking from all the related sources (yellow highlight): http://genome.ucsc.edu/s/brianlee/mm10_chrM
This explains how there can be a discrepancy between the number of positions covered by the phastCons file and the length of a chromosome. It is likely caused by gaps in the multiple alignment process. Gaps often result from gaps in the reference assembly or variable regions that do not align well between species. Anywhere the multiple alignment is lacking data, phastCons cannot be used to calculated conservation scores.
Author: brianlee
Session Name: mm10_chrM
Genome Assembly: mm10
Creation Date: 2019-11-22
Views: 2199
|
1574409600 |
2199 |
 |
Description: Juniata Bioinformatics KH JL
Lethal neonatal rigidity and multifocal seizure syndrome is characterized by underdeveloped brains and seizures that begin in-utero. Seizures continue after birth and result in death which occurs usually by the age of 4 months. The disease is caused by an insertion of an A in the BRAT1 gene. The normal BRAT1 gene function is to be a master controller of cell cycled checkpoint signaling pathways that are required for cellular responses to DNA damage.
Author: herrkx18
Session Name: Amish Lethal Neonatal rigidity and Seizure syndrome
Genome Assembly: hg19
Creation Date: 2020-02-10
Views: 1552
|
1581321600 |
1552 |
 |
Description:
Author: tacazares
Session Name: maxATAC ATAC-seq Data
Genome Assembly: hg38
Creation Date: 2022-04-11
Views: 795
|
1649664000 |
795 |
 |
Description:
Author: Erinija
Session Name: NA12878 WES Benchmark
Genome Assembly: hg19
Creation Date: 2019-11-28
Views: 2720
|
1574928000 |
2720 |
 |
Description:
Author: mia_21
Session Name: mitochonrial cyb mutation
Genome Assembly: hg38
Creation Date: 2019-10-31
Views: 1673
|
1572508800 |
1673 |
 |
Description:
Author: Brigitte
Session Name: snp hla vph
Genome Assembly: hg38
Creation Date: 2019-10-24
Views: 1642
|
1571904000 |
1642 |
 |
Description:
Author: cocopow
Session Name: hg38_genes24336
Genome Assembly: hg38
Creation Date: 2019-10-23
Views: 2339
|
1571817600 |
2339 |
 |
Description:
Author: cocopow
Session Name: hg19_LINC00299
Genome Assembly: hg19
Creation Date: 2019-10-25
Views: 1716
|
1571990400 |
1716 |
 |
Description:
Author: sguc
Session Name: miR-122_KO_genotyping_primers
Genome Assembly: mm10
Creation Date: 2019-10-17
Views: 1753
|
1571299200 |
1753 |
 |
Description:
Author: ani_davis
Session Name: TAS2R38 group6
Genome Assembly: hg19
Creation Date: 2022-02-01
Views: 800
|
1643702400 |
800 |
 |
Description:
Author: zhangh357
Session Name: hg19_m6A_site
Genome Assembly: hg19
Creation Date: 2019-10-14
Views: 1882
|
1571040000 |
1882 |
 |
Description:
Author: cocopow
Session Name: dm6_transcripts
Genome Assembly: dm6
Creation Date: 2019-10-09
Views: 1733
|
1570608000 |
1733 |
 |
Description:
Author: ykumar84
Session Name: mm10_CRE
Genome Assembly: mm10
Creation Date: 2019-10-07
Views: 1794
|
1570435200 |
1794 |
 |
Description:
Author: ykumar84
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2019-10-04
Views: 1984
|
1570176000 |
1984 |
 |
Description:
Author: mecay.1
Session Name: session 1
Genome Assembly: hg38
Creation Date: 2020-02-10
Views: 1847
|
1581321600 |
1847 |
 |
Description:
Author: cocopow
Session Name: hg38_ENCreg
Genome Assembly: hg38
Creation Date: 2019-10-03
Views: 1708
|
1570089600 |
1708 |
 |
Description:
Author: cocopow
Session Name: hg38_hg19lo_24241C
Genome Assembly: hg38
Creation Date: 2019-10-02
Views: 2071
|
1570003200 |
2071 |
 |
Description:
Author: cocopow
Session Name: hg38_hg19lo_24241B
Genome Assembly: hg38
Creation Date: 2019-10-02
Views: 1937
|
1570003200 |
1937 |
 |
Description:
Author: cocopow
Session Name: hg38_hg19lo_24241A
Genome Assembly: hg38
Creation Date: 2019-10-02
Views: 1807
|
1570003200 |
1807 |
 |
Description:
Author: sallen
Session Name: DSP-hg38
Genome Assembly: hg38
Creation Date: 2020-02-11
Views: 1665
|
1581408000 |
1665 |
 |
Description:
Author: rgmilian
Session Name: hg19_BamHIsite
Genome Assembly: hg19
Creation Date: 2019-09-25
Views: 910
|
1569398400 |
910 |
 |
Description:
Author: rgmilian
Session Name: hg19_20p13_exon_view_flaggedSNPs+exons
Genome Assembly: hg19
Creation Date: 2019-09-24
Views: 799
|
1569312000 |
799 |
 |
Description:
Author: barloweml
Session Name: YeastPracticeMLB
Genome Assembly: sacCer3
Creation Date: 2019-09-24
Views: 1781
|
1569312000 |
1781 |
 |
Description:
Author: ykumar84
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2019-09-18
Views: 2183
|
1568793600 |
2183 |
 |
Description:
Author: IsaacGelman
Session Name: mm10_h3k4me3
Genome Assembly: mm10
Creation Date: 2019-11-01
Views: 1651
|
1572595200 |
1651 |
 |
Description:
Author: nheyer
Session Name: New_wiggles
Genome Assembly: hg38
Creation Date: 2019-11-12
Views: 1631
|
1573545600 |
1631 |
 |
Description: CircRNA annotation as identified in the publication "Evolutionarily young transposable elements drive circular RNA repertoires" by Gruhl et al. (in preparation). The browser view include an additional track with the repeat dimers surrounding the respective circRNA.
Author: Frenzchen
Session Name: hg38 circRNA annotation
Genome Assembly: hg38
Creation Date: 2019-02-10
Views: 1536
|
1549785600 |
1536 |
 |
Description: CircRNA and parental gene annotation as identified in the publication "Evolutionarily young transposable elements drive circular RNA repertoires" by Gruhl et al. (in preparation). The browser view include an additional track with the repeat dimers surrounding the respective circRNA.
Author: Frenzchen
Session Name: rn5 circRNA annotation
Genome Assembly: rn5
Creation Date: 2016-09-23
Views: 1561
|
1474617600 |
1561 |
 |
Description: CircRNA and parental gene annotation as identified in the publication "Evolutionarily young transposable elements drive circular RNA repertoires" by Gruhl et al. (in preparation). The browser view include an additional track with the repeat dimers surrounding the respective circRNA.
Author: Frenzchen
Session Name: mm10 circRNA annotation
Genome Assembly: mm10
Creation Date: 2016-09-23
Views: 1522
|
1474617600 |
1522 |
 |
Description: CircRNA and parental gene annotation as identified in the publication "Evolutionarily young transposable elements drive circular RNA repertoires" by Gruhl et al. (in preparation). The browser view include an additional track with the repeat dimers surrounding the respective circRNA.
Author: Frenzchen
Session Name: monDom5 circRNA annotation
Genome Assembly: monDom5
Creation Date: 2016-09-23
Views: 1584
|
1474617600 |
1584 |
 |
Description:
Author: MarcoDS
Session Name: B_to_PSC_reprogramming
Genome Assembly: mm10
Creation Date: 2019-09-16
Views: 1673
|
1568620800 |
1673 |
 |
Description:
Author: bethpay
Session Name: mm10_il7r_findenhancerpromoter
Genome Assembly: mm10
Creation Date: 2019-09-09
Views: 1861
|
1568016000 |
1861 |
 |
Description:
Author: rafal.czapiewski@ed.ac.uk
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2019-09-09
Views: 1546
|
1568016000 |
1546 |
Screenshot not available
Click Here to view |
Description:
Author: tolvayha
Session Name: Fall18 Worm test HT
Genome Assembly: ce11
Creation Date: 2018-08-30
Views: 1835
|
1535616000 |
1835 |
Screenshot not available
Click Here to view |
Description:
Author: tolvayha
Session Name: RHO - Photoreceptor
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1785
|
1542096000 |
1785 |
 |
Description:
Author: swarnaseetha
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2019-10-23
Views: 1655
|
1571817600 |
1655 |
 |
Description: ReMap 2018: An atlas of regulatory regions from an integrative analysis of Human DNA-binding sequencing experiments. Go to <a href="http://remap.univ-amu.fr">ReMap</a> for more info. <a href="https://academic.oup.com/nar/article/46/D1/D267/4602873">ReMap 2018 paper</a>.
Author: Benoit Ballester
Session Name: US_hg38_ReMap2018
Genome Assembly: hg38
Creation Date: 2017-08-11
Views: 3396
|
1502438400 |
3396 |
 |
Description:
Author: rasou2ba
Session Name: hg19 - rs12357257 - Rasoul
Genome Assembly: hg19
Creation Date: 2017-10-31
Views: 2005
|
1509436800 |
2005 |
 |
Description:
Author: pesinojv
Session Name: TAS2R38 group#4
Genome Assembly: hg19
Creation Date: 2018-01-22
Views: 1855
|
1516608000 |
1855 |
 |
Description:
Author: oshgo
Session Name: 14q32Methyl450-GO8
Genome Assembly: hg19
Creation Date: 2015-10-19
Views: 2159
|
1445241600 |
2159 |
 |
Description: GH01J209814 is an enhancer associated with the gene IRF6 (Interferon Regulatory Factor 6). Coding mutations in IRF6 are known to be pathogenic, causing diseases related to cleft lip and cleft palate, such as Van Der Woude Syndrome 1 (VWS1). GH01J209814, an enhancer of IRF6 located ~10kb upstream from the gene, harbors regulatory non-coding variants strongly associated with VWS1.
Author: simonf
Session Name: IRF6 cleft lip enhancer
Genome Assembly: hg19
Creation Date: 2018-11-28
Views: 1865
|
1543392000 |
1865 |
 |
Description:
Author: australia2018
Session Name: hg18_EP_bamSnps_custom
Genome Assembly: hg18
Creation Date: 2018-11-07
Views: 1848
|
1541577600 |
1848 |
 |
Description:
Author: rafal.czapiewski@ed.ac.uk
Session Name: BAC and FOSMID
Genome Assembly: mm10
Creation Date: 2019-03-27
Views: 1819
|
1553673600 |
1819 |
 |
Description:
Author: jazmynperez
Session Name: APP
Genome Assembly: hg19
Creation Date: 2018-11-07
Views: 2522
|
1541577600 |
2522 |
 |
Description:
Author: huijing
Session Name: check_pacbio
Genome Assembly: dm6
Creation Date: 2018-11-06
Views: 1064
|
1541491200 |
1064 |
 |
Description:
Author: cehnouz
Session Name: test1
Genome Assembly: hg19
Creation Date: 2017-08-25
Views: 1977
|
1503648000 |
1977 |
 |
Description:
Author: jtm
Session Name: shiny_app_hg38_v2
Genome Assembly: hg38
Creation Date: 2018-11-05
Views: 3075
|
1541404800 |
3075 |
 |
Description:
Author: ruhollah
Session Name: 18_chr17
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1965
|
1541059200 |
1965 |
 |
Description:
Author: ruhollah
Session Name: 20_chr1
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1868
|
1541059200 |
1868 |
 |
Description:
Author: ruhollah
Session Name: 14_chr2
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 2107
|
1541059200 |
2107 |
 |
Description:
Author: ruhollah
Session Name: 17_chr3
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1891
|
1541059200 |
1891 |
 |
Description:
Author: ruhollah
Session Name: 19_chr10
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1836
|
1541059200 |
1836 |
 |
Description:
Author: ruhollah
Session Name: 16_chr5
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1823
|
1541059200 |
1823 |
 |
Description:
Author: ruhollah
Session Name: 6_chr11
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1836
|
1541059200 |
1836 |
 |
Description:
Author: ruhollah
Session Name: 8_chr2
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 2139
|
1541059200 |
2139 |
 |
Description:
Author: ruhollah
Session Name: 11_chr10
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1811
|
1541059200 |
1811 |
 |
Description:
Author: ruhollah
Session Name: 12_chr3
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 2120
|
1541059200 |
2120 |
 |
Description:
Author: ruhollah
Session Name: 13_chr3
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1848
|
1541059200 |
1848 |
 |
Description:
Author: ruhollah
Session Name: 15_chr19
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1773
|
1541059200 |
1773 |
 |
Description:
Author: ruhollah
Session Name: 7_chr11
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 2130
|
1541059200 |
2130 |
 |
Description:
Author: ruhollah
Session Name: 9_chr20
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1817
|
1541059200 |
1817 |
 |
Description:
Author: ruhollah
Session Name: 5_chr16
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1934
|
1541059200 |
1934 |
 |
Description:
Author: ruhollah
Session Name: 4_chr3
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1807
|
1541059200 |
1807 |
 |
Description:
Author: ruhollah
Session Name: 10_chr17
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1808
|
1541059200 |
1808 |
 |
Description:
Author: ruhollah
Session Name: 3_chr14
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1817
|
1541059200 |
1817 |
 |
Description:
Author: ruhollah
Session Name: 2_chr8
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1943
|
1541059200 |
1943 |
 |
Description:
Author: ruhollah
Session Name: 1_chr15
Genome Assembly: hg38
Creation Date: 2018-11-01
Views: 1756
|
1541059200 |
1756 |
 |
Description: This session highlights data from the DASHR v2.0 Hub, created by the Wang lab at the University of Pennsylvania. DASHR is a comprehensive database of expression and processing information to date on all major classes of human small non-coding RNA (sncRNA) genes and mature sncNA annotations, expression levels, sequence and RNA processing information across 185 human tissues, cell types, and cell lines.
The region being visualized shows mir-132. A sncRNA linked to post-transcriptional regulation of neuronal cells.
More info on DASHR V2.0: http://dashr2.lisanwanglab.org/
Author: Lou
Session Name: DASHRV2
Genome Assembly: hg38
Creation Date: 2018-12-11
Views: 2092
|
1544515200 |
2092 |
 |
Description:
Author: sharma.896
Session Name: TP53
Genome Assembly: hg19
Creation Date: 2018-10-30
Views: 2063
|
1540886400 |
2063 |
 |
Description: human p53 ChIP-seq coverage depth (tracks) & SISSRs peak calls with associated differential gene expression from published data as of May 2015
Author: cocapo
Session Name: human p53 Binding And Expression Resource (BAER) hub
Genome Assembly: hg19
Creation Date: 2018-10-30
Views: 2156
|
1540886400 |
2156 |
 |
Description:
Author: cocopow
Session Name: hg19_rn6_geneHancer
Genome Assembly: hg19
Creation Date: 2019-01-18
Views: 2031
|
1547798400 |
2031 |
 |
Description:
Author: cocopow
Session Name: hg38_GeneHancer
Genome Assembly: hg38
Creation Date: 2019-01-28
Views: 1869
|
1548662400 |
1869 |
 |
Description:
Author: apacis
Session Name: Wu_ARMC5_ChIP_C3G
Genome Assembly: mm10
Creation Date: 2020-10-08
Views: 1253
|
1602144000 |
1253 |
 |
Description: This sessionView is a collection of tracks centered around large genetic variants (CNVs). The session is organized with gene annotations at the top, followed by the <a href="https://academic.oup.com/nar/article/42/D1/D986/1068860" target="_blank">DGV Struct Var track</a>, which displays variants observed in healthy individuals. The rest of the tracks include different databases that visualize large variants linked to disease phenotypes. The region displayed is a 2Mbp region on chr4 with many pathological variants present.
Author: view
Session Name: VariationCNVs
Genome Assembly: hg19
Creation Date: 2018-10-16
Views: 1973
|
1539676800 |
1973 |
 |
Description: This sessionView is a collection of tracks centered around genetic variation. The collection of tracks primarily displays small variants from multiple databases. The session is organized with tracks having fewer variants with more annotations near the top, and large tracks with fewer annotations near the bottom. The region displayed is the <a href="https://ghr.nlm.nih.gov/gene/SOD1" target="_blank">SOD1 gene</a> which is well annotated and provides an exemplar for this track collection.
Author: view
Session Name: Variation
Genome Assembly: hg19
Creation Date: 2018-10-10
Views: 1865
|
1539158400 |
1865 |
 |
Description: This sessionView is a collection of tracks centered around comparative genomics. It primarily consists of multiz alignments and conserved elements/conservation scores identified by <a href="http://compgen.cshl.edu/phast/" target="_blank">phastCons</a>. 18 organisms were chosen for comparison from 100 available vertebrates, sampling clades evenly. The region displayed is an ultra-conserved region in the PCBP2 gene where little divergence is seen throughout all mammals. A similar session is available, <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=ConservationDiv" target="_blank">ConservationDiv</a>, which looks at a highly divergent region of the genome.
Author: view
Session Name: ConservationCons
Genome Assembly: hg19
Creation Date: 2018-10-10
Views: 2818
|
1539158400 |
2818 |
 |
Description: This sessionView is a collection of tracks centered around comparative genomics. It primarily consists of multiz alignments and conserved elements/conservation scores identified by <a href="http://compgen.cshl.edu/phast/" target="_blank">phastCons</a>. 18 organisms were chosen for comparison from 100 available vertebrates, sampling clades evenly. The region displayed is a <a href="https://www.hhmi.org/news/researchers-identify-human-dna-fast-track" target="_blank">human accelerated region (HAR)</a>. HARs are among the most divergent regions in the genome displaying high divergence even among primates. An alternate region of this session is also available, <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=ConservationCons" target="_blank">ConservationCons</a>, which looks at a highly conserved region of the genome.
Author: view
Session Name: ConservationDiv
Genome Assembly: hg19
Creation Date: 2018-10-10
Views: 2770
|
1539158400 |
2770 |
 |
Description: This sessionView is a collection of tracks centered around clinical significance. The track composition is similar to the <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=Clinical" target="_blank">Clinical</a> public session, however, the number of tracks has been reduced to increase clarity. Featured tracks include gene annotations, SNVs, CNVs, SVs, and our publications track built by mining sequences and SNPs in publications. The displayed region is focused around the HTT gene, responsible for Huntington's disease and Lopes-Maciel-Rodan syndrome. The large region, which also includes part of the adjacent GRK4 gene, showcases the more concise view of this track in contrast to the <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=Clinical" target="_blank">Clinical</a> session. A similar session is also available which shows the bp resolution CAG repeat linked to Huntington's disease, <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=ClinicalZoom" target="_blank">ClinicalZoom</a>.
Author: view
Session Name: ClinicalLite
Genome Assembly: hg19
Creation Date: 2018-10-10
Views: 3158
|
1539158400 |
3158 |
 |
Description: This sessionView is a collection of tracks centered around gene support. The region displayed includes the <a href="https://www.ncbi.nlm.nih.gov/gene /3198" target="_blank"> HOXA1</a> gene and its upstream neighbor <a href="https://www.ncbi.nlm.nih.gov/gene/100506311" target="_blank"> HOTAIRM1</a>. The view is organized with different gene annotation approaches at the top, followed by mRNA evidence, and TSS data at the bottom.
Author: view
Session Name: GeneSupport
Genome Assembly: hg19
Creation Date: 2018-10-05
Views: 3228
|
1538726400 |
3228 |
 |
Description:
Author: xingjie
Session Name: FMR1-AFF2-iPSC screening
Genome Assembly: hg38
Creation Date: 2018-10-05
Views: 2055
|
1538726400 |
2055 |
 |
Description:
Author: EsterCastillo
Session Name: VanesaGarcia_AS
Genome Assembly: hg38
Creation Date: 2018-08-14
Views: 2115
|
1534233600 |
2115 |
 |
Description:
Author: EsterCastillo
Session Name: SandraHernandez_UPF
Genome Assembly: hg38
Creation Date: 2018-12-10
Views: 1747
|
1544428800 |
1747 |
 |
Description:
Author: EsterCastillo
Session Name: NuriaSala_ICO_AS
Genome Assembly: hg19
Creation Date: 2018-06-18
Views: 1801
|
1529308800 |
1801 |
 |
Description:
Author: EsterCastillo
Session Name: NRCC_HospTxagorritxo_Guiomar
Genome Assembly: hg19
Creation Date: 2018-08-01
Views: 1839
|
1533110400 |
1839 |
 |
Description:
Author: EsterCastillo
Session Name: Meme_ROIs_AmpliSeq
Genome Assembly: hg19
Creation Date: 2018-12-18
Views: 1765
|
1545120000 |
1765 |
 |
Description:
Author: EsterCastillo
Session Name: MarcSorigue
Genome Assembly: hg19
Creation Date: 2018-08-24
Views: 2232
|
1535097600 |
2232 |
 |
Description:
Author: EsterCastillo
Session Name: LifeSequencing_JuanMartinez
Genome Assembly: hg19
Creation Date: 2018-05-18
Views: 1816
|
1526630400 |
1816 |
 |
Description:
Author: EsterCastillo
Session Name: LauraMuinelo_CHUS_AS
Genome Assembly: hg38
Creation Date: 2018-05-31
Views: 2007
|
1527753600 |
2007 |
 |
Description:
Author: EsterCastillo
Session Name: HURyC_Gloria_Francisco_25bp_padding
Genome Assembly: hg19
Creation Date: 2019-03-24
Views: 2462
|
1553414400 |
2462 |
 |
Description:
Author: EsterCastillo
Session Name: HURyC_Gloria_Francisco_10bp_padding
Genome Assembly: hg19
Creation Date: 2019-03-24
Views: 1698
|
1553414400 |
1698 |
 |
Description:
Author: EsterCastillo
Session Name: HDR_LuciaPerezCarbonero_HospBurgos_NMs
Genome Assembly: hg19
Creation Date: 2018-09-06
Views: 1795
|
1536220800 |
1795 |
 |
Description:
Author: EsterCastillo
Session Name: HDR_LuciaPerezCarbonero_HospBurgos
Genome Assembly: hg19
Creation Date: 2018-08-24
Views: 1765
|
1535097600 |
1765 |
 |
Description:
Author: EsterCastillo
Session Name: Genycell_FJD_Tcell
Genome Assembly: hg19
Creation Date: 2019-05-28
Views: 1652
|
1559030400 |
1652 |
 |
Description:
Author: EsterCastillo
Session Name: Genycell_FJD_Bcell
Genome Assembly: hg19
Creation Date: 2019-05-28
Views: 1682
|
1559030400 |
1682 |
 |
Description:
Author: EsterCastillo
Session Name: BarbaraTazon_AS_Myeloid
Genome Assembly: hg19
Creation Date: 2018-04-09
Views: 1875
|
1523260800 |
1875 |
 |
Description:
Author: EsterCastillo
Session Name: Arnau_DrViladell_AmpliSeq
Genome Assembly: hg19
Creation Date: 2019-05-08
Views: 1610
|
1557302400 |
1610 |
 |
Description:
Author: EsterCastillo
Session Name: AnaSebio_HospStPau
Genome Assembly: hg19
Creation Date: 2018-05-01
Views: 2000
|
1525161600 |
2000 |
 |
Description:
Author: EsterCastillo
Session Name: AnaOsorio_AS_CNIO
Genome Assembly: hg19
Creation Date: 2018-05-24
Views: 1873
|
1527148800 |
1873 |
 |
Description:
Author: EsterCastillo
Session Name: AmpliSeq_Meme_Concierge2
Genome Assembly: hg19
Creation Date: 2019-02-11
Views: 1727
|
1549872000 |
1727 |
 |
Description:
Author: EsterCastillo
Session Name: AmpliSeq_Meme_Concierge
Genome Assembly: hg19
Creation Date: 2019-01-31
Views: 1798
|
1548921600 |
1798 |
 |
Description:
Author: EsterCastillo
Session Name: AmpliSeq Focus
Genome Assembly: hg19
Creation Date: 2018-08-20
Views: 1755
|
1534752000 |
1755 |
 |
Description:
Author: EsterCastillo
Session Name: AmpliSeq Cancer HotSpots v2
Genome Assembly: hg19
Creation Date: 2018-06-06
Views: 1718
|
1528272000 |
1718 |
 |
Description:
Author: EsterCastillo
Session Name: 12Oct_todos37g_DLW
Genome Assembly: hg19
Creation Date: 2019-01-20
Views: 2178
|
1547971200 |
2178 |
 |
Description:
Author: tzaro
Session Name: mm9
Genome Assembly: mm9
Creation Date: 2018-10-08
Views: 2105
|
1538985600 |
2105 |
 |
Description:
Author: zheyangshan
Session Name: hg19 GRO-seq EGFR shjun
Genome Assembly: hg19
Creation Date: 2019-01-16
Views: 1631
|
1547625600 |
1631 |
 |
Description:
Author: sajjeev
Session Name: sj39_40
Genome Assembly: hg19
Creation Date: 2019-01-17
Views: 2348
|
1547712000 |
2348 |
 |
Description:
Author: cocopow
Session Name: mm10_limbs
Genome Assembly: mm10
Creation Date: 2019-01-15
Views: 1956
|
1547539200 |
1956 |
 |
Description:
Author: emmawentworth
Session Name: Adult-Mouse-Adipose
Genome Assembly: mm10
Creation Date: 2019-04-29
Views: 1779
|
1556524800 |
1779 |
 |
Description:
Author: Heywhoyou22
Session Name: AMD SNP C9 2
Genome Assembly: hg19
Creation Date: 2018-10-02
Views: 1802
|
1538467200 |
1802 |
 |
Description:
Author: brittatj
Session Name: Fall19 Yeast test(TB)
Genome Assembly: sacCer3
Creation Date: 2019-08-27
Views: 1575
|
1566892800 |
1575 |
 |
Description:
Author: Matteo
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2018-10-02
Views: 1758
|
1538467200 |
1758 |
 |
Description:
Author: cocapo
Session Name: dense2pack
Genome Assembly: hg38
Creation Date: 2018-09-26
Views: 1915
|
1537948800 |
1915 |
 |
Description:
Author: akhoke
Session Name: KMT2E/SRPK2_2
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1945
|
1537862400 |
1945 |
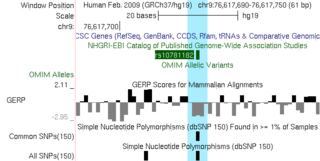 |
Description:
Author: mazawm
Session Name: hg19#19SingleNucleotideResolution
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1575
|
1537862400 |
1575 |
 |
Description:
Author: benbeijs
Session Name: AMD SNP (JSB), CFH Exercise 2 (SNPs)
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1877
|
1537862400 |
1877 |
 |
Description:
Author: anglemem
Session Name: AMD SNP 2
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1925
|
1537862400 |
1925 |
 |
Description:
Author: akhoke
Session Name: KMT2E/SRPK2
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1882
|
1537862400 |
1882 |
 |
Description:
Author: benbeijs
Session Name: AMD SNP (JSB), CFH Exercise 1
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1892
|
1537862400 |
1892 |
 |
Description:
Author: mazawm
Session Name: hg19#19GenomicContext
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1628
|
1537862400 |
1628 |
 |
Description:
Author: Heywhoyou22
Session Name: AMD SNP C9 zoom
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1717
|
1537862400 |
1717 |
 |
Description:
Author: Heywhoyou22
Session Name: AMD SNP C9
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1752
|
1537862400 |
1752 |
 |
Description:
Author: Heywhoyou22
Session Name: HGD
Genome Assembly: hg19
Creation Date: 2018-09-25
Views: 1808
|
1537862400 |
1808 |
 |
Description:
Author: mazawm
Session Name: hg19HGDmazawm2
Genome Assembly: hg19
Creation Date: 2018-09-19
Views: 1780
|
1537344000 |
1780 |
 |
Description:
Author: akhoke
Session Name: HGD2
Genome Assembly: hg19
Creation Date: 2018-09-18
Views: 1786
|
1537257600 |
1786 |
 |
Description:
Author: benbeijs
Session Name: UCSC/ApE (JSB) Exercise, Exon 10
Genome Assembly: hg19
Creation Date: 2018-09-18
Views: 1745
|
1537257600 |
1745 |
 |
Description:
Author: white4cj
Session Name: Fall 19 Yeast Test (JW, PA, MG)
Genome Assembly: sacCer3
Creation Date: 2019-08-27
Views: 1631
|
1566892800 |
1631 |
 |
Description:
Author: akhoke
Session Name: HGD
Genome Assembly: hg19
Creation Date: 2018-09-18
Views: 1790
|
1537257600 |
1790 |
 |
Description:
Author: mazawm
Session Name: hg19HGDmazawm
Genome Assembly: hg19
Creation Date: 2018-09-18
Views: 1857
|
1537257600 |
1857 |
 |
Description:
Author: anglemem
Session Name: HGD Gene (EMA)
Genome Assembly: hg19
Creation Date: 2018-09-18
Views: 1841
|
1537257600 |
1841 |
 |
Description:
Author: benbeijs
Session Name: UCSC/ApE (JSB) Exercise
Genome Assembly: hg19
Creation Date: 2018-09-18
Views: 1718
|
1537257600 |
1718 |
 |
Description:
Author: yangyangyh
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2018-09-16
Views: 1781
|
1537084800 |
1781 |
 |
Description: The LCT gene, responsible for digesting lactose, and the nearby MCM6 gene, which harbors variants that affect the expression of LCT.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_LCTandMCM6
Genome Assembly: hg19
Creation Date: 2022-06-22
Views: 869
|
1655884800 |
869 |
 |
Description:
Author: dcaetan2
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2017-12-05
Views: 2104
|
1512460800 |
2104 |
 |
Description:
Author: cocopow
Session Name: hg19_chrM
Genome Assembly: hg19
Creation Date: 2019-08-13
Views: 2429
|
1565683200 |
2429 |
 |
Description:
Author: ruifang
Session Name: TAC3
Genome Assembly: hg38
Creation Date: 2019-08-11
Views: 1656
|
1565510400 |
1656 |
 |
Description:
Author: cocopow
Session Name: panTro6_refSeq
Genome Assembly: panTro6
Creation Date: 2019-08-20
Views: 1671
|
1566288000 |
1671 |
 |
Description:
Author: Alexandra Despang
Session Name: PWD_SNPS_mm10
Genome Assembly: mm10
Creation Date: 2016-01-15
Views: 1596
|
1452844800 |
1596 |
 |
Description:
Author: mziegler
Session Name: encode_Accessibility _4
Genome Assembly: hg19
Creation Date: 2018-09-06
Views: 1945
|
1536220800 |
1945 |
 |
Description:
Author: mziegler
Session Name: encode_Accessibility _3
Genome Assembly: hg19
Creation Date: 2018-09-06
Views: 1950
|
1536220800 |
1950 |
 |
Description:
Author: mziegler
Session Name: encode_Accessibility _2
Genome Assembly: hg19
Creation Date: 2018-09-06
Views: 1951
|
1536220800 |
1951 |
 |
Description:
Author: mazawm
Session Name: hg19withDSPandMYBPC1
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1797
|
1536048000 |
1797 |
 |
Description:
Author: akhoke
Session Name: hg19_9/4/18
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1802
|
1536048000 |
1802 |
 |
Description:
Author: Heywhoyou22
Session Name: DSP
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1889
|
1536048000 |
1889 |
 |
Description:
Author: Heywhoyou22
Session Name: MYBPC1
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1774
|
1536048000 |
1774 |
 |
Description:
Author: zheyangshan
Session Name: hg19 BCL2
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1691
|
1536048000 |
1691 |
 |
Description:
Author: abby.stapleton
Session Name: Spring17 Mouse Test
Genome Assembly: mm9
Creation Date: 2019-07-29
Views: 1789
|
1564387200 |
1789 |
 |
Description:
Author: abby.stapleton
Session Name: Spring17 Human Test
Genome Assembly: hg38
Creation Date: 2019-07-29
Views: 1567
|
1564387200 |
1567 |
 |
Description:
Author: abby.stapleton
Session Name: Spring17 Worm Test
Genome Assembly: ce11
Creation Date: 2019-07-29
Views: 1569
|
1564387200 |
1569 |
 |
Description:
Author: abby.stapleton
Session Name: Spring17 Yeast Test
Genome Assembly: sacCer3
Creation Date: 2019-07-29
Views: 1587
|
1564387200 |
1587 |
 |
Description:
Author: jwjiao
Session Name: PRDM16KO1_E15WT1.anno
Genome Assembly: hg38
Creation Date: 2019-08-05
Views: 1601
|
1564992000 |
1601 |
 |
Description:
Author: xgwZRyfWLidWxULv
Session Name: RPE_public
Genome Assembly: hg19
Creation Date: 2019-01-15
Views: 1657
|
1547539200 |
1657 |
 |
Description:
Author: elencampoy
Session Name: HsInv0379
Genome Assembly: hg19
Creation Date: 2021-02-25
Views: 1092
|
1614240000 |
1092 |
 |
Description:
Author: bondurlc
Session Name: TAS2R38 group 3
Genome Assembly: hg19
Creation Date: 2018-09-03
Views: 1793
|
1535961600 |
1793 |
 |
Description:
Author: ruhollah
Session Name: hg19_118Chr22_nmt4
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1755
|
1547452800 |
1755 |
 |
Description:
Author: ruhollah
Session Name: hg19_100Chr19_nmt3
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1714
|
1547452800 |
1714 |
 |
Description:
Author: mazawm
Session Name: mazawm/hg19
Genome Assembly: hg19
Creation Date: 2018-08-31
Views: 1777
|
1535702400 |
1777 |
 |
Description:
Author: erhuoyi
Session Name: suva
Genome Assembly: hg38
Creation Date: 2018-08-31
Views: 1849
|
1535702400 |
1849 |
 |
Description:
Author: benbeijs
Session Name: MYBPC1 (JSB) Assignment
Genome Assembly: hg19
Creation Date: 2018-08-30
Views: 2206
|
1535616000 |
2206 |
 |
Description:
Author: akhoke
Session Name: Human Genome DSP
Genome Assembly: hg19
Creation Date: 2018-08-30
Views: 1768
|
1535616000 |
1768 |
 |
Description:
Author: anglemem
Session Name: EmilyAnglemyer 8/30 In class browser assignment
Genome Assembly: hg19
Creation Date: 2018-08-30
Views: 1794
|
1535616000 |
1794 |
 |
Description:
Author: Morgan_T
Session Name: Aug30 Asignment MT
Genome Assembly: hg19
Creation Date: 2018-08-30
Views: 1776
|
1535616000 |
1776 |
 |
Description:
Author: stjacqrm
Session Name: Spring18_DSP_RMSJ
Genome Assembly: hg19
Creation Date: 2018-08-30
Views: 1899
|
1535616000 |
1899 |
 |
Description:
Author: benbeijs
Session Name: CGEMS Worm (JSB) Test
Genome Assembly: ce11
Creation Date: 2018-08-30
Views: 1848
|
1535616000 |
1848 |
 |
Description:
Author: sschuma
Session Name: Fall18 DSP (SS)
Genome Assembly: hg19
Creation Date: 2018-08-30
Views: 1734
|
1535616000 |
1734 |
 |
Description:
Author: akhoke
Session Name: Yeast AKH
Genome Assembly: sacCer3
Creation Date: 2018-08-30
Views: 1993
|
1535616000 |
1993 |
 |
Description:
Author: erick.gabriel
Session Name: ICD-1 gene (by Erick)
Genome Assembly: ce11
Creation Date: 2018-08-30
Views: 1842
|
1535616000 |
1842 |
 |
Description:
Author: mazawm
Session Name: mazawm8/30
Genome Assembly: ce11
Creation Date: 2018-08-30
Views: 1925
|
1535616000 |
1925 |
 |
Description:
Author: akhoke
Session Name: CGEMS yeast AKH test
Genome Assembly: sacCer3
Creation Date: 2018-08-30
Views: 1798
|
1535616000 |
1798 |
 |
Description:
Author: Morgan_T
Session Name: Fall18 Yeast test M.T.
Genome Assembly: sacCer3
Creation Date: 2018-08-30
Views: 1751
|
1535616000 |
1751 |
 |
Description:
Author: foxaa
Session Name: Fall18 Worm test AF
Genome Assembly: ce11
Creation Date: 2018-08-30
Views: 2352
|
1535616000 |
2352 |
 |
Description:
Author: Heywhoyou22
Session Name: Spring18 Worm test MRK
Genome Assembly: ce11
Creation Date: 2018-08-30
Views: 1760
|
1535616000 |
1760 |
 |
Description:
Author: stjacqrm
Session Name: Spring18_Yeast_test_RMSJ
Genome Assembly: sacCer3
Creation Date: 2018-08-30
Views: 1992
|
1535616000 |
1992 |
 |
Description:
Author: hammer
Session Name: piranha-eclip_sclip2
Genome Assembly: hg38
Creation Date: 2018-08-30
Views: 1894
|
1535616000 |
1894 |
 |
Description:
Author: sschuma
Session Name: Fall18 Yeast Test (SS)
Genome Assembly: sacCer3
Creation Date: 2018-08-30
Views: 1753
|
1535616000 |
1753 |
 |
Description:
Author: Heywhoyou22
Session Name: forenke2
Genome Assembly: hg38
Creation Date: 2018-11-29
Views: 1849
|
1543478400 |
1849 |
 |
Description:
Author: Heywhoyou22
Session Name: forenke
Genome Assembly: hg38
Creation Date: 2018-11-29
Views: 1793
|
1543478400 |
1793 |
 |
Description:
Author: Boonede
Session Name: TAS2R38#6
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1633
|
1547452800 |
1633 |
 |
Description: RYiu-90010430-MEDG580-UBC-2020
Author: cpddoraemon
Session Name: RYiu-90010430-MEDG580-UBC-2020
Genome Assembly: hg38
Creation Date: 2022-10-09
Views: 666
|
1665302400 |
666 |
 |
Description:
Author: anzelm
Session Name: hg19-hrc3
Genome Assembly: hg19
Creation Date: 2018-03-19
Views: 2075
|
1521446400 |
2075 |
 |
Description:
Author: anzelm
Session Name: hg19-180-10-3
Genome Assembly: hg19
Creation Date: 2018-03-19
Views: 1827
|
1521446400 |
1827 |
 |
Description:
Author: anglemem
Session Name: MYBPC1 Anglemyer
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1738
|
1536048000 |
1738 |
 |
Description:
Author: anglemem
Session Name: DSP
Genome Assembly: hg19
Creation Date: 2018-09-05
Views: 2161
|
1536134400 |
2161 |
 |
Description:
Author: anglemem
Session Name: 10/2 custom track assignment
Genome Assembly: hg19
Creation Date: 2018-10-02
Views: 1991
|
1538467200 |
1991 |
 |
Description:
Author: allisontaggart
Session Name: hg19_BPs
Genome Assembly: hg19
Creation Date: 2017-01-18
Views: 2287
|
1484726400 |
2287 |
 |
Description:
Author: alexc
Session Name: CytoSettings
Genome Assembly: hg19
Creation Date: 2018-03-13
Views: 2139
|
1520928000 |
2139 |
 |
Description:
Author: alexaabdelaziz
Session Name: colored_clusters_Alexa_AA
Genome Assembly: hg19
Creation Date: 2017-02-24
Views: 2092
|
1487923200 |
2092 |
 |
Description:
Author: akhoke
Session Name: MYBPC1
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1815
|
1536048000 |
1815 |
 |
Description:
Author: akhoke
Session Name: DSP
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1849
|
1536048000 |
1849 |
 |
Description:
Author: akhoke
Session Name: CRLs3
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1788
|
1542096000 |
1788 |
 |
Description:
Author: akhoke
Session Name: CRLs2
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1762
|
1542096000 |
1762 |
 |
Description:
Author: akhoke
Session Name: CRLs
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1769
|
1542096000 |
1769 |
 |
Description:
Author: akhoke
Session Name: AMD SNP Custom Track
Genome Assembly: hg19
Creation Date: 2018-10-03
Views: 1795
|
1538553600 |
1795 |
 |
Description:
Author: airibarren.1
Session Name: Ventana buena + Hubs
Genome Assembly: hg38
Creation Date: 2018-02-06
Views: 1730
|
1517904000 |
1730 |
 |
Description:
Author: airibarren.1
Session Name: Regulatory elements; epigentic marks
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 2228
|
1519632000 |
2228 |
 |
Description:
Author: airibarren.1
Session Name: Oregano
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1956
|
1519632000 |
1956 |
 |
Description:
Author: airibarren.1
Session Name: NHLF
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1929
|
1519632000 |
1929 |
 |
Description:
Author: airibarren.1
Session Name: NHEK
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1896
|
1519632000 |
1896 |
 |
Description:
Author: airibarren.1
Session Name: K562
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1993
|
1519632000 |
1993 |
 |
Description:
Author: airibarren.1
Session Name: HUVEC
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 2248
|
1519632000 |
2248 |
 |
Description:
Author: airibarren.1
Session Name: HSMM
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 2064
|
1519632000 |
2064 |
 |
Description:
Author: airibarren.1
Session Name: H1-hESC
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 2147
|
1519632000 |
2147 |
 |
Description:
Author: airibarren.1
Session Name: Genes and gene expression
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1932
|
1519632000 |
1932 |
 |
Description:
Author: airibarren.1
Session Name: GM12878
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1930
|
1519632000 |
1930 |
 |
Description:
Author: airibarren.1
Session Name: Evolution
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 2084
|
1519632000 |
2084 |
 |
Description:
Author: airibarren.1
Session Name: Ensembl segment
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1892
|
1519632000 |
1892 |
 |
Description:
Author: airibarren.1
Session Name: Ensembl activity
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1923
|
1519632000 |
1923 |
 |
Description:
Author: airibarren.1
Session Name: ChIp
Genome Assembly: hg19
Creation Date: 2018-02-26
Views: 1944
|
1519632000 |
1944 |
 |
Description:
Author: agacita
Session Name: Regulatory_Variants_Heart_333
Genome Assembly: hg19
Creation Date: 2018-05-11
Views: 2008
|
1526025600 |
2008 |
 |
Description:
Author: agacita
Session Name: Regulatory_Var_89912022_labels
Genome Assembly: hg19
Creation Date: 2018-05-10
Views: 2446
|
1525939200 |
2446 |
 |
Description:
Author: agacita
Session Name: Regulatory_Var_89912022_Final
Genome Assembly: hg19
Creation Date: 2018-05-11
Views: 2031
|
1526025600 |
2031 |
 |
Description:
Author: agacita
Session Name: Regulatory_Var_89912022
Genome Assembly: hg19
Creation Date: 2018-05-10
Views: 1793
|
1525939200 |
1793 |
 |
Description:
Author: afanas
Session Name: hg19-H3K27Ac
Genome Assembly: hg19
Creation Date: 2017-11-27
Views: 2346
|
1511769600 |
2346 |
 |
Description:
Author: gerbermm
Session Name: TAS2R38 Grp6 MG
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 2047
|
1547452800 |
2047 |
 |
Description:
Author: hammer
Session Name: piranha-eclip_sclip
Genome Assembly: hg38
Creation Date: 2018-08-27
Views: 1842
|
1535356800 |
1842 |
 |
Description:
Author: hkao
Session Name: SMARCC1 4-3
Genome Assembly: hg38
Creation Date: 2019-07-09
Views: 1595
|
1562659200 |
1595 |
 |
Description:
Author: hammer
Session Name: ablife
Genome Assembly: hg38
Creation Date: 2018-08-27
Views: 1918
|
1535356800 |
1918 |
 |
Description: RNA-seq in cell lines were carried out as follows: after MGC803 cells were transfected with siRNA against circMRPS35 or corresponding control in triplicate, total RNA were extracted and cDNA libraries were constructed for rRNA-depleted sample using the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina according to the manufacturer’s instructions, and were sequenced on Illumina HiSeq Xten. RPKM method was used to determine the gene expression. The raw data were analyzed according to the DESeq2 method. Pathway analysis was used to find out the significant pathway of the differential genes according to KEGG database. For gene ontology (GO) analysis, we downloaded the GO annotations from NCBI, UniProt and the Gene Ontology. Fisher’s exact test was applied to identify the significant GO categories and FDR was used to correct the p-values.
Author: jieMM
Session Name: hg38_FINAL
Genome Assembly: hg38
Creation Date: 2019-11-05
Views: 1986
|
1572940800 |
1986 |
 |
Description:
Author: leonic
Session Name: bing li et al. m6A RIP
Genome Assembly: mm10
Creation Date: 2018-08-21
Views: 1867
|
1534838400 |
1867 |
 |
Description:
Author: zxtzhangqian
Session Name: 3a-140-IDH
Genome Assembly: mm9
Creation Date: 2018-08-20
Views: 2095
|
1534752000 |
2095 |
 |
Description:
Author: davis2018
Session Name: hg19_example_asl
Genome Assembly: hg19
Creation Date: 2018-08-20
Views: 1803
|
1534752000 |
1803 |
 |
Description:
Author: ruhollah
Session Name: hg19_29ChrX_nmt23
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1882
|
1543219200 |
1882 |
 |
Description:
Author: ruhollah
Session Name: hg19_13Chr3_nmt23
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1700
|
1543219200 |
1700 |
 |
Description: This session shows how the multi-region tool allows you to swap in alternate sequences into the main sequence, when desired. For example, if you click the "mutli-region" button under the Browser image (or go to the top "View" menu and then select it), you can paste chr5_KI270794v1_alt in the "Show one alternate haplotype, placed on its chromosome, using ID:" box and then also click the " Highlight alternating regions in multi-region view" box and the result is this session.<br> <br> This issue came up when a user was doing PCR and found two results, one on chr5 and one on chr5_KI270794v1_alt and wanted to understand the meaning of the chr5_..._alt PCR hit. On our gateway page for hg38 (http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg38) you will find an Assembly Details section, which will explain how the Genome Reference Consortium (GRC) in building a reference assembly observed several human chromosomal regions exhibit sufficient variability to prevent adequate representation by a single sequence and to address this issue, the GRCh38/hg38 assembly provides alternate sequence for selected variant regions through the inclusion of alternate loci scaffolds (or alt loci). The assembly contains 261 alt loci, where this chr5_KI270794v1_alt is one of the regions that just happens to be right around the specific gene of interest. This GRC Assembly Terminology page is useful in learning the definition of alternate sequences: https://www.ncbi.nlm.nih.gov/grc/help/definitions/ The answer to the user was that in some ways one can interpret this additional chr5_KI270794v1_alt PCR result as a representation where the GRC determined an additional sequence provides an alternate representation of the same locus. And this session helps to visualize that alternate sequence in place around the rest of the chromosome.
<img src="https://groups.google.com/a/soe.ucsc.edu/group/genome/attach/d6cb8d335b5b/image.png?part=0.1&authuser=0" alt="PCR results">
Author: brianlee
Session Name: hg38_ chr5_KI270794v1_alt
Genome Assembly: hg38
Creation Date: 2018-08-10
Views: 1819
|
1533888000 |
1819 |
 |
Description:
Author: chubukovp
Session Name: hg19_SMC_upload
Genome Assembly: hg19
Creation Date: 2016-10-14
Views: 2170
|
1476432000 |
2170 |
 |
Description:
Author: anglemem
Session Name: CGEMS Worm EA Test
Genome Assembly: ce11
Creation Date: 2018-08-30
Views: 1742
|
1535616000 |
1742 |
 |
Description:
Author: ruhollah
Session Name: hg19_170Chr1_nmt4
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1694
|
1547452800 |
1694 |
 |
Description:
Author: ruhollah
Session Name: hg19_173Chr2_nmt4
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1708
|
1547452800 |
1708 |
 |
Description: RNA-seq data from Lee HC et al. Open Biol. 2020 Feb. Molecular anatomy of the pre-primitive-streak chick embryo.
Author: stern_lab
Session Name: Lee HC et al. chick pre-primitive streak
Genome Assembly: galGal5
Creation Date: 2020-06-25
Views: 1616
|
1593072000 |
1616 |
 |
Description:
Author: cocopow
Session Name: hg38_bw_GSM1131536
Genome Assembly: hg38
Creation Date: 2019-03-07
Views: 1675
|
1551945600 |
1675 |
 |
Description:
Author: cocopow
Session Name: hg38_HGMDvsCurated
Genome Assembly: hg38
Creation Date: 2019-03-06
Views: 1915
|
1551859200 |
1915 |
 |
Description:
Author: cocopow
Session Name: danRer11_bw_GSM1131536
Genome Assembly: danRer11
Creation Date: 2019-03-07
Views: 1601
|
1551945600 |
1601 |
 |
Description:
Author: cocopow
Session Name: mm10_bejTB
Genome Assembly: mm10
Creation Date: 2019-08-01
Views: 1572
|
1564646400 |
1572 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chrX
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1799
|
1532505600 |
1799 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr22
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1788
|
1532505600 |
1788 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr17
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1856
|
1532505600 |
1856 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr16
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1754
|
1532505600 |
1754 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr14
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1747
|
1532505600 |
1747 |
 |
Description:
Author: Gabriel
Session Name: LDTF-Kras
Genome Assembly: mm10
Creation Date: 2020-03-26
Views: 1533
|
1585209600 |
1533 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr18
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1667
|
1532505600 |
1667 |
 |
Description:
Author: dorothyzhao
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2019-08-13
Views: 1563
|
1565683200 |
1563 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr19
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1731
|
1532505600 |
1731 |
 |
Description: ReMap 2020: An atlas of regulatory regions from an integrative analysis Arabidopsis thaliana DNA-binding sequencing experiments. Go to <a href="http://remap.univ-amu.fr">ReMap</a> for more info.
Author: Benoit Ballester
Session Name: US_ReMap2020_Thaliana
Genome Assembly: hub_1936559_araTha1
Creation Date: 2019-08-29
Views: 10037
|
1567065600 |
10037 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr15
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1836
|
1532505600 |
1836 |
 |
Description:
Author: dantaki
Session Name: sv2_merge_debug
Genome Assembly: hg19
Creation Date: 2018-11-08
Views: 1809
|
1541664000 |
1809 |
 |
Description:
Author: dantaki
Session Name: ccr_hg19
Genome Assembly: hg19
Creation Date: 2018-12-12
Views: 1814
|
1544601600 |
1814 |
 |
Description: this is a test
Author: dandan_0909
Session Name: hg19_4_17
Genome Assembly: hg19
Creation Date: 2018-04-15
Views: 157
|
1523779200 |
157 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr6
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1944
|
1532505600 |
1944 |
 |
Description:
Author: danrlu
Session Name: Fuller_paper
Genome Assembly: dm6
Creation Date: 2019-10-25
Views: 2084
|
1571990400 |
2084 |
 |
Description:
Author: mersedehr
Session Name: NSUN4_v2_rs66575205
Genome Assembly: hg19
Creation Date: 2018-06-15
Views: 1770
|
1529049600 |
1770 |
 |
Description: This sessionView is a collection of tracks centered around epigenomics. The profiles displayed primarily belong to the H1 cell line, however different origin samples may be chosen in the track configurations. The display is organized with gene annotations and mRNA abundance first, followed by regulatory profiles in the form of <a href="https://www.ncbi.nlm.nih.gov/pubmed/21441907" target="_blank">chromatin states (ChromHMM)</a>. These data are followed by ChIP-seq tracks exploring various transcription factors and regulatory regions/promoter tracks looking at DNase/CpG/methylation. The final tracks are the <a href="https://www.nature.com/articles/ng.2653" target="_blank">GTEx combined eQTL</a>, which identifies genetic variants likely affecting proximal gene expression, and the GTEx Gene Expression, which shows median gene expression levels in 51 tissues and 2 cell lines. The region visualized is a ~12,000bp window surrounding the <a href="https://ghr.nlm.nih.gov/gene/BRCA1" target="_blank">BRCA1 gene</a> transcription start site. <a href="https://ghr.nlm.nih.gov/gene/BRCA1" target="_blank">BRCA1</a> is a tumor suppressor and housekeeping gene linked to increased susceptibility to breast cancer when <a href="https://www.ncbi.nlm.nih.gov/pubmed/16846527" target="_blank">certain epigenomic markers are present</a>.
Author: view
Session Name: Epigenomics
Genome Assembly: hg19
Creation Date: 2018-10-10
Views: 1886
|
1539158400 |
1886 |
 |
Description: This sessionView is a collection of tracks centered around clinical significance. Featured tracks include gene annotations, SNVs, CNVs, SVs, and our publications track built by mining sequences and SNPs in publications. The displayed region is a 294bp window looking at the CAG repeat linked to Huntington’s disease in the HTT gene. Two similar sessions are also available: <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=ClinicalLite" target="_blank">ClinicalLite</a> which has a reduced number of tracks for increased clarity, and <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=Clinical" target="_blank">Clinical</a> which is the most informative clinical view.
Author: view
Session Name: ClinicalZoom
Genome Assembly: hg19
Creation Date: 2018-10-10
Views: 13413
|
1539158400 |
13413 |
 |
Description: This sessionView is a collection of tracks centered around clinical significance. Featured tracks include gene annotations, SNVs, CNVs, SVs, a cancer mutation database and our publications track built by mining sequences and SNPs in publications. The displayed region is focused around the HTT gene, responsible for Huntington's disease and Lopes-Maciel-Rodan syndrome. Two similar sessions are also available: <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=ClinicalZoom" target="_blank">ClinicalZoom</a> which shows the bp resolution CAG repeat linked to Huntington's disease, as well as <a href="http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=view&hgS_otherUserSessionName=ClinicalLite" target="_blank">ClinicalLite</a> which has a reduced number of tracks for increased clarity.
Author: view
Session Name: Clinical
Genome Assembly: hg19
Creation Date: 2018-10-10
Views: 3579
|
1539158400 |
3579 |
 |
Description: This sessionView is a collection of tracks centered around assembly support. The region displayed includes a reported GRC incident on hg19 which was then patched. It also showcases a gap which can lead to inconsistent or missing data on existing tracks; as seen by the Mappability and 1000 Genomes Project Accessible Regions tracks.
Author: view
Session Name: AssemblySupport
Genome Assembly: hg19
Creation Date: 2018-10-05
Views: 1855
|
1538726400 |
1855 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr13
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1975
|
1532505600 |
1975 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr12
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1802
|
1532505600 |
1802 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr10
Genome Assembly: hg19
Creation Date: 2018-07-24
Views: 2246
|
1532419200 |
2246 |
 |
Description:
Author: csitans
Session Name: Mouse ncRNA HSCs
Genome Assembly: mm9
Creation Date: 2017-06-19
Views: 1834
|
1497859200 |
1834 |
 |
Description:
Author: cschee
Session Name: RNA_HEMa
Genome Assembly: hg19
Creation Date: 2016-06-14
Views: 2170
|
1465891200 |
2170 |
 |
Description:
Author: cschee
Session Name: RNA_A375
Genome Assembly: hg19
Creation Date: 2016-06-14
Views: 2037
|
1465891200 |
2037 |
 |
Description:
Author: cschee
Session Name: A375_K27me3
Genome Assembly: hg19
Creation Date: 2016-06-14
Views: 2024
|
1465891200 |
2024 |
 |
Description:
Author: cschee
Session Name: A375_K27Ac
Genome Assembly: hg19
Creation Date: 2016-06-15
Views: 2040
|
1465977600 |
2040 |
 |
Description:
Author: cschee
Session Name: A375_Input_21092016
Genome Assembly: hg19
Creation Date: 2016-09-21
Views: 2205
|
1474444800 |
2205 |
 |
Description:
Author: cschee
Session Name: A375_Input
Genome Assembly: hg19
Creation Date: 2016-06-15
Views: 1975
|
1465977600 |
1975 |
 |
Description:
Author: cosmid88
Session Name: SRY-SOX9-ChIP-Chip
Genome Assembly: mm8
Creation Date: 2013-06-29
Views: 2553
|
1372492800 |
2553 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr9
Genome Assembly: hg19
Creation Date: 2018-07-24
Views: 1713
|
1532419200 |
1713 |
 |
Description:
Author: cyoung
Session Name: Young, Fall17, Uba1 example
Genome Assembly: mm9
Creation Date: 2017-09-10
Views: 1787
|
1505030400 |
1787 |
 |
Description:
Author: cyoung
Session Name: Fall17 Bio630 CY test
Genome Assembly: mm9
Creation Date: 2017-09-13
Views: 1765
|
1505289600 |
1765 |
 |
Description:
Author: cyoung
Session Name: Bio630 CRX variants Young
Genome Assembly: hg38
Creation Date: 2017-09-26
Views: 1828
|
1506412800 |
1828 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr8
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1760
|
1532505600 |
1760 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr7
Genome Assembly: hg19
Creation Date: 2018-07-24
Views: 1717
|
1532419200 |
1717 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr5
Genome Assembly: hg19
Creation Date: 2018-07-24
Views: 2046
|
1532419200 |
2046 |
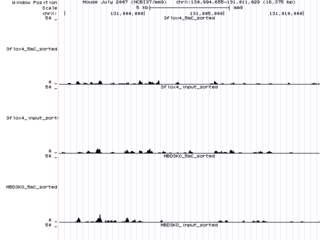 |
Description:
Author: ddunican
Session Name: MBD3 WT v KO medip-seq ion_torrent mm9
Genome Assembly: mm9
Creation Date: 2016-09-26
Views: 2198
|
1474876800 |
2198 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr4
Genome Assembly: hg19
Creation Date: 2018-07-24
Views: 1906
|
1532419200 |
1906 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr3
Genome Assembly: hg19
Creation Date: 2018-07-24
Views: 1810
|
1532419200 |
1810 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr2
Genome Assembly: hg19
Creation Date: 2018-07-24
Views: 2264
|
1532419200 |
2264 |
 |
Description:
Author: Welekie
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2020-05-20
Views: 1444
|
1589961600 |
1444 |
 |
Description:
Author: simonwang612
Session Name: worm_ChIP_seq_L4_0716
Genome Assembly: ce10
Creation Date: 2018-07-16
Views: 1871
|
1531728000 |
1871 |
 |
Description:
Author: olena
Session Name: OTC-regulatory
Genome Assembly: hg19
Creation Date: 2018-07-13
Views: 1995
|
1531468800 |
1995 |
 |
Description:
Author: jtm
Session Name: shiny_app_mm10_v1
Genome Assembly: mm10
Creation Date: 2018-07-11
Views: 2949
|
1531296000 |
2949 |
 |
Description:
Author: jtm
Session Name: shiny_app_hg38_v1
Genome Assembly: hg38
Creation Date: 2018-07-11
Views: 5057
|
1531296000 |
5057 |
 |
Description:
Author: Kyle@Arkell_Lab
Session Name: hg38_ZIC2_3'UTR_RNA_Annotation
Genome Assembly: hg38
Creation Date: 2018-07-10
Views: 1999
|
1531209600 |
1999 |
 |
Description:
Author: tonyzeng
Session Name: apa
Genome Assembly: hg19
Creation Date: 2018-07-06
Views: 2060
|
1530864000 |
2060 |
 |
Description:
Author: speese
Session Name: dm6 FlyVar track
Genome Assembly: dm6
Creation Date: 2018-07-05
Views: 1581
|
1530777600 |
1581 |
 |
Description:
Author: Mouro_81
Session Name: B6J vs 129S1 SNPs at Lrrfip2/Mlh1
Genome Assembly: mm10
Creation Date: 2017-05-15
Views: 2008
|
1494835200 |
2008 |
 |
Description:
Author: MeganDurham
Session Name: BRAF
Genome Assembly: hg19
Creation Date: 2018-04-24
Views: 1971
|
1524556800 |
1971 |
 |
Description:
Author: Leif.Benner
Session Name: Leif_session
Genome Assembly: dm3
Creation Date: 2015-04-11
Views: 1985
|
1428739200 |
1985 |
 |
Description:
Author: Lab252
Session Name: Roche_et_al_NAR_44_19_9315_2016
Genome Assembly: hg19
Creation Date: 2016-07-04
Views: 2114
|
1467619200 |
2114 |
 |
Description:
Author: Kurotaki
Session Name: mm10 tachibana
Genome Assembly: mm10
Creation Date: 2016-05-12
Views: 2094
|
1463040000 |
2094 |
 |
Description:
Author: Ketty
Session Name: hg38_ChiPseq CRISPR clones
Genome Assembly: hg38
Creation Date: 2019-06-20
Views: 1651
|
1561017600 |
1651 |
 |
Description:
Author: KateLawrensonCSHS
Session Name: OCAC_CIMBA_Oncoarray
Genome Assembly: hg19
Creation Date: 2016-05-03
Views: 2656
|
1462262400 |
2656 |
 |
Description:
Author: Vizoso1
Session Name: UBP10
Genome Assembly: sacCer3
Creation Date: 2017-07-25
Views: 2085
|
1500969600 |
2085 |
 |
Description:
Author: Vizoso1
Session Name: RSA4
Genome Assembly: sacCer3
Creation Date: 2017-07-25
Views: 2160
|
1500969600 |
2160 |
 |
Description:
Author: Vizoso1
Session Name: NMD3
Genome Assembly: sacCer3
Creation Date: 2017-07-25
Views: 1930
|
1500969600 |
1930 |
 |
Description:
Author: Vizoso1
Session Name: ENP1
Genome Assembly: sacCer3
Creation Date: 2017-07-25
Views: 2026
|
1500969600 |
2026 |
 |
Description:
Author: Tung
Session Name: ndst1_10snp
Genome Assembly: hg38
Creation Date: 2018-04-13
Views: 2106
|
1523606400 |
2106 |
Screenshot not available
Click Here to view |
Description:
Author: Toma Matveeva
Session Name: CGEMS #1
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 2072
|
1469088000 |
2072 |
Screenshot not available
Click Here to view |
Description:
Author: Tao Zhang
Session Name: chicken lncRNA
Genome Assembly: hub_30425_galGal4
Creation Date: 2016-11-28
Views: 2317
|
1480320000 |
2317 |
 |
Description:
Author: Robin
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2016-10-04
Views: 2063
|
1475568000 |
2063 |
 |
Description:
Author: Robin
Session Name: CHPv3 versus Sanger/CHPv2
Genome Assembly: hg19
Creation Date: 2016-07-11
Views: 1971
|
1468224000 |
1971 |
 |
Description:
Author: Rahman.team
Session Name: CART38A
Genome Assembly: hg38
Creation Date: 2018-10-30
Views: 1896
|
1540886400 |
1896 |
 |
Description:
Author: Rahman.team
Session Name: CART37A
Genome Assembly: hg19
Creation Date: 2018-10-30
Views: 1959
|
1540886400 |
1959 |
 |
Description:
Author: cocopow
Session Name: hg38_23676
Genome Assembly: hg38
Creation Date: 2019-06-20
Views: 1562
|
1561017600 |
1562 |
 |
Description:
Author: cocopow
Session Name: hg19_superDuperTrack
Genome Assembly: hg19
Creation Date: 2019-04-16
Views: 1877
|
1555401600 |
1877 |
 |
Description:
Author: cocopow
Session Name: hg19_DMR_chr2
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1658
|
1554105600 |
1658 |
 |
Description:
Author: cocopow
Session Name: hg19_DMR_1chr1_P180_P21_exm_3
Genome Assembly: hg19
Creation Date: 2019-03-21
Views: 1611
|
1553155200 |
1611 |
 |
Description:
Author: cocopow
Session Name: hg19_ChIA
Genome Assembly: hg19
Creation Date: 2019-03-21
Views: 1649
|
1553155200 |
1649 |
 |
Description:
Author: cocopow
Session Name: hg19_BedGraph
Genome Assembly: hg19
Creation Date: 2019-03-29
Views: 1730
|
1553846400 |
1730 |
 |
Description:
Author: cocopow
Session Name: danRer7_bw_GSM1131536
Genome Assembly: danRer7
Creation Date: 2019-03-07
Views: 1655
|
1551945600 |
1655 |
 |
Description:
Author: britnyblu
Session Name: rena_atac_data
Genome Assembly: mm10
Creation Date: 2019-08-15
Views: 1588
|
1565856000 |
1588 |
 |
Description:
Author: megallegos
Session Name: Biol 310: Module 2 TSS-seq
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 5942
|
1536048000 |
5942 |
 |
Description:
Author: mjulialamberti
Session Name: cxcl12
Genome Assembly: hg38
Creation Date: 2019-08-08
Views: 1605
|
1565251200 |
1605 |
 |
Description:
Author: vperreau@unimelb.edu.au
Session Name: mm10 Linnarsson Celltype autoscale
Genome Assembly: mm10
Creation Date: 2019-02-18
Views: 948
|
1550476800 |
948 |
 |
Description:
Author: masuping
Session Name: chr_lncRNA-seq_CPM
Genome Assembly: hg19
Creation Date: 2019-10-14
Views: 1550
|
1571040000 |
1550 |
 |
Description: Sept 10, 2024
Author: rchelsea
Session Name: hg38 CRX
Genome Assembly: hg38
Creation Date: 2024-09-11
Views: 221
|
1726041600 |
221 |
 |
Description:
Author: sajjeev
Session Name: foxa1_07222019
Genome Assembly: hg19
Creation Date: 2019-07-22
Views: 1602
|
1563782400 |
1602 |
Screenshot not available
Click Here to view |
Description:
Author: sajjeev
Session Name: ICR_cmyc_07222019
Genome Assembly: hg19
Creation Date: 2019-07-22
Views: 1625
|
1563782400 |
1625 |
Screenshot not available
Click Here to view |
Description:
Author: sajjeev
Session Name: 07222019
Genome Assembly: hg19
Creation Date: 2019-07-22
Views: 1541
|
1563782400 |
1541 |
 |
Description:
Author: mauriege
Session Name: Fall21 Yeast test (GEM)
Genome Assembly: sacCer3
Creation Date: 2021-09-04
Views: 979
|
1630742400 |
979 |
 |
Description:
Author: celiagonzalez98
Session Name: Class 1
Genome Assembly: hg38
Creation Date: 2020-02-10
Views: 1460
|
1581321600 |
1460 |
 |
Description:
Author: batra.anjali
Session Name: Spring19 Yeast test AB
Genome Assembly: sacCer3
Creation Date: 2019-01-11
Views: 1649
|
1547193600 |
1649 |
 |
Description:
Author: ruhollah
Session Name: hg19_1025_chr5_nmtf14
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1554
|
1554105600 |
1554 |
 |
Description:
Author: ruhollah
Session Name: hg19_830_chr6_nmtf15
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1583
|
1554105600 |
1583 |
 |
Description:
Author: ruhollah
Session Name: hg19_750_chr19_nmtf15
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1571
|
1554105600 |
1571 |
 |
Description:
Author: ruhollah
Session Name: hg19_199_chr8_nmtf16
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1589
|
1554105600 |
1589 |
 |
Description:
Author: ruhollah
Session Name: hg19_1405_chr16_nmtf16
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1890
|
1554105600 |
1890 |
 |
Description:
Author: ruhollah
Session Name: hg19_212_chr17_nmtf16
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1690
|
1554105600 |
1690 |
 |
Description:
Author: ruhollah
Session Name: hg19_131_chr2_nmtf17
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1607
|
1554105600 |
1607 |
 |
Description:
Author: ruhollah
Session Name: hg19_1163_chr1_nmtf18
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1649
|
1554105600 |
1649 |
 |
Description:
Author: ruhollah
Session Name: hg19_414_Chr8_nmtf19
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1621
|
1554105600 |
1621 |
 |
Description:
Author: ruhollah
Session Name: hg19_1617_chr1_nmtf20
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1635
|
1554105600 |
1635 |
 |
Description:
Author: ruhollah
Session Name: hg19_795_chr19_nmtf23
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1643
|
1554105600 |
1643 |
 |
Description:
Author: Boody09
Session Name: Boudreau Lab_human cardiac Ago2 HITS-CLIP
Genome Assembly: hg19
Creation Date: 2018-05-08
Views: 2026
|
1525766400 |
2026 |
 |
Description:
Author: ntinamu001
Session Name: NCOMMS-19-03087;HCT116_vs_DKO1
Genome Assembly: hg19
Creation Date: 2019-08-22
Views: 1546
|
1566460800 |
1546 |
 |
Description:
Author: ntinamu001
Session Name: NCOMMS-19-03087;Mouse_OE
Genome Assembly: mm9
Creation Date: 2019-08-22
Views: 1362
|
1566460800 |
1362 |
 |
Description:
Author: ntinamu001
Session Name: NCOMMS-19-03087;SH-SY5Y
Genome Assembly: hg19
Creation Date: 2019-08-22
Views: 1429
|
1566460800 |
1429 |
 |
Description:
Author: AlicePsyche
Session Name: zebrafish_enhancers
Genome Assembly: danRer7
Creation Date: 2018-04-04
Views: 1785
|
1522828800 |
1785 |
 |
Description:
Author: tschauer
Session Name: Harpprecht_NAR
Genome Assembly: dm6
Creation Date: 2019-03-27
Views: 1682
|
1553673600 |
1682 |
 |
Description:
Author: Boonede
Session Name: Spring19 Yeast Test DB
Genome Assembly: sacCer3
Creation Date: 2019-01-11
Views: 1694
|
1547193600 |
1694 |
 |
Description:
Author: frongemg
Session Name: Spring19 Yeast test MGF
Genome Assembly: sacCer3
Creation Date: 2019-01-11
Views: 1634
|
1547193600 |
1634 |
 |
Description:
Author: ruhollah
Session Name: hg19_172Chr3_nmt8
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1818
|
1547452800 |
1818 |
 |
Description:
Author: ruckerhr
Session Name: Spring19 Yeast HR
Genome Assembly: sacCer3
Creation Date: 2019-01-09
Views: 1681
|
1547020800 |
1681 |
 |
Description:
Author: ruckerhr
Session Name: TAS2R8group4
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1809
|
1547452800 |
1809 |
 |
Description:
Author: Artem Babaian
Session Name: LIONS_ENCODE_supplement
Genome Assembly: hg19
Creation Date: 2019-01-09
Views: 1739
|
1547020800 |
1739 |
 |
Description:
Author: gerbermm
Session Name: Spring19 Yeast test MG
Genome Assembly: sacCer3
Creation Date: 2019-01-08
Views: 1740
|
1546934400 |
1740 |
 |
Description:
Author: descentj
Session Name: Spring19YeastTestTD
Genome Assembly: sacCer3
Creation Date: 2019-01-08
Views: 1794
|
1546934400 |
1794 |
 |
Description:
Author: shashipujar
Session Name: Multiz_conservation_SP
Genome Assembly: hg38
Creation Date: 2018-06-18
Views: 1954
|
1529308800 |
1954 |
 |
Description:
Author: agacita
Session Name: DYB_Promoter
Genome Assembly: hg19
Creation Date: 2019-01-22
Views: 1738
|
1548144000 |
1738 |
 |
Description:
Author: lbenner
Session Name: fs(1)K741
Genome Assembly: dm6
Creation Date: 2017-09-11
Views: 1933
|
1505116800 |
1933 |
 |
Description:
Author: mersedehr
Session Name: NSUN4_v1_rs72886903
Genome Assembly: hg19
Creation Date: 2018-06-14
Views: 1692
|
1528963200 |
1692 |
 |
Description:
Author: shashaverygood@163.com
Session Name: DGRP2snp
Genome Assembly: dm3
Creation Date: 2018-06-14
Views: 1676
|
1528963200 |
1676 |
 |
Description:
Author: Annaneed
Session Name: Anna_GEL
Genome Assembly: mm9
Creation Date: 2018-06-08
Views: 1414
|
1528444800 |
1414 |
Screenshot not available
Click Here to view |
Description:
Author: view
Session Name: 11
Genome Assembly: hg38
Creation Date: 2018-12-21
Views: 1750
|
1545379200 |
1750 |
 |
Description:
Author: GDJ_Duke
Session Name: Layden_liver_ATACseq
Genome Assembly: mm10
Creation Date: 2018-12-16
Views: 1710
|
1544947200 |
1710 |
 |
Description:
Author: nedbrewer66
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2018-12-18
Views: 1867
|
1545120000 |
1867 |
 |
Description:
Author: mfarkas03
Session Name: 1299To p53 mutants
Genome Assembly: hg19
Creation Date: 2018-06-07
Views: 1307
|
1528358400 |
1307 |
 |
Description:
Author: arem
Session Name: hg19-cc
Genome Assembly: hg19
Creation Date: 2019-02-08
Views: 2037
|
1549612800 |
2037 |
 |
Description:
Author: arem
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2019-02-08
Views: 2183
|
1549612800 |
2183 |
 |
Description:
Author: Mikhailova_li
Session Name: KB-1471A8.1
Genome Assembly: hg19
Creation Date: 2018-06-04
Views: 1976
|
1528099200 |
1976 |
 |
Description:
Author: Mikhailova_li
Session Name: session 3
Genome Assembly: hg19
Creation Date: 2018-06-03
Views: 1733
|
1528012800 |
1733 |
 |
Description:
Author: Mikhailova_li
Session Name: session 1
Genome Assembly: hg19
Creation Date: 2018-06-03
Views: 1742
|
1528012800 |
1742 |
 |
Description:
Author: brianlee
Session Name: hg38_chrM
Genome Assembly: hg38
Creation Date: 2018-06-01
Views: 1821
|
1527840000 |
1821 |
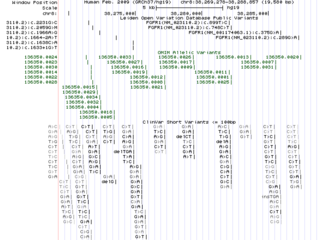 |
Description: Our LOVD Variants track has variants from many LSDBs; our OMIM Alleles, GWAS Catalog, ClinVar Variants and other tracks in the Phenotypes & Literature group may provide clues to when a SNP is Clinical (LSDB,OMIM,TPA,Diagnostic)" per the Flagged SNPs track based on whether dbSNP's true-or-false "clinically-assoc" flag is set from NCBI's documentation.
Author: brianlee
Session Name: hg19_variants
Genome Assembly: hg19
Creation Date: 2018-06-01
Views: 1955
|
1527840000 |
1955 |
 |
Description:
Author: nehoralevi
Session Name: CTCF- macs2 control2
Genome Assembly: mm10
Creation Date: 2019-06-10
Views: 1583
|
1560153600 |
1583 |
 |
Description: Epigenome map of human fetal and iPSC-derived retinal pigment epithelium (RPE)
Author: s041629
Session Name: iPSCORE_RPE_session_hg19
Genome Assembly: hg19
Creation Date: 2018-05-31
Views: 2201
|
1527753600 |
2201 |
 |
Description: This session highlights data from the Brain Epigenome Hub, created by Peter Hickey, Lindsay Rizzardi, and Kaspar Hansen and others at Johns Hopkins University. The hub shows methylation, ATAC-seq, and RNAseq across different brain regions.
Author: chmalee
Session Name: hg19_brainEpigenome
Genome Assembly: hg19
Creation Date: 2018-05-30
Views: 2943
|
1527667200 |
2943 |
 |
Description:
Author: rezo
Session Name: ReCappable-seq
Genome Assembly: hg38
Creation Date: 2019-08-11
Views: 8000
|
1565510400 |
8000 |
 |
Description: 6 CLEAR-CLIP tracks from mouse keratinocytes in total. 3 showing CLEAR-CLIP reads from all miRNAs and 3 showing only miR-200 family specific reads. For each set of three there are Controls, miR-200 DKO and miR-200 inducible libraries.
Author: Bjerkega
Session Name: Bjerkega CLEAR-CLIP
Genome Assembly: mm10
Creation Date: 2019-10-04
Views: 1723
|
1570176000 |
1723 |
 |
Description:
Author: ltoker
Session Name: Marzi
Genome Assembly: hg19
Creation Date: 2021-01-05
Views: 1449
|
1609833600 |
1449 |
 |
Description:
Author: carlchancc
Session Name: 20191017_MC_Forelimb_Hindlimb_hg19
Genome Assembly: hg19
Creation Date: 2019-10-17
Views: 1670
|
1571299200 |
1670 |
 |
Description:
Author: hkao
Session Name: RHOA 2-2
Genome Assembly: hg38
Creation Date: 2019-07-09
Views: 1519
|
1562659200 |
1519 |
 |
Description:
Author: hkao
Session Name: NEFM 1-3
Genome Assembly: hg38
Creation Date: 2019-07-09
Views: 1564
|
1562659200 |
1564 |
 |
Description:
Author: ejlopezsoto
Session Name: Cacna1b e35-e41
Genome Assembly: hg19
Creation Date: 2019-07-11
Views: 1744
|
1562832000 |
1744 |
 |
Description: All patient isolates in PRJNA613958, with exception of MinION sequenced, were individually aligned against the NC_045512.2 reference with SNPs called for each isolate using 'ngskit4b kalign'. A matrix of all individual isolate SNP site loci (rows) and isolate base calls at that loci (cols) created with 'ngskit4b snpmarkers' and this matrix then processed with 'ngskit4b snps2pgsnps' to output the pgSNPs tracks. Tracks show, for each SNP loci, the number of isolates containing the displayed allele base. Tracks provided for age, sex and sample month.
Author: biodiscovery
Session Name: wuhCor1 PRJNA613958 allele components
Genome Assembly: wuhCor1
Creation Date: 2020-04-29
Views: 1625
|
1588147200 |
1625 |
 |
Description:
Author: eldadshulman
Session Name: Homer_newaproch
Genome Assembly: mm10
Creation Date: 2019-06-30
Views: 1515
|
1561881600 |
1515 |
 |
Description:
Author: ruhollah
Session Name: hg19_162Chr19_nmt4
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1625
|
1547452800 |
1625 |
 |
Description:
Author: 704855238
Session Name: chip-seq_LiuZX_seh1
Genome Assembly: rn6
Creation Date: 2018-05-24
Views: 1881
|
1527148800 |
1881 |
 |
Description:
Author: zhangr100
Session Name: AHBEC_VitD
Genome Assembly: hg19
Creation Date: 2019-04-04
Views: 1685
|
1554364800 |
1685 |
 |
Description:
Author: liqing9102
Session Name: mm9
Genome Assembly: mm9
Creation Date: 2018-05-16
Views: 1821
|
1526457600 |
1821 |
 |
Description:
Author: avlasova
Session Name: tADseq.short-library.all_frames
Genome Assembly: hub_271523_TFbb_list180_linear
Creation Date: 2018-05-14
Views: 1941
|
1526284800 |
1941 |
 |
Description: Juniata Bioinformatics KH JL
Lethal neonatal rigidity and multifocal seizure syndrome is characterized by underdeveloped brains and seizures that begin in-utero. Seizures continue after birth and result in death which occurs usually by the age of 4 months. The disease is caused by an insertion of an A in the BRAT1 gene. The normal BRAT1 gene function is to be a master controller of cell cycled checkpoint signaling pathways that are required for cellular responses to DNA damage.
Author: herrkx18
Session Name: Amish Lethal Neonatal rigidity and Seizure syndrome 2
Genome Assembly: hg19
Creation Date: 2020-02-27
Views: 1414
|
1582790400 |
1414 |
 |
Description:
Author: Vaa2001
Session Name: hg19_ASXL1Gen
Genome Assembly: hg38
Creation Date: 2019-10-24
Views: 1485
|
1571904000 |
1485 |
 |
Description: one fly one genome: H3 hybrid fly, hybrid genome sequencing and assembly
Author: msadams
Session Name: H3-2_dm6
Genome Assembly: dm6
Creation Date: 2019-10-24
Views: 632
|
1571904000 |
632 |
Screenshot not available
Click Here to view |
Description:
Author: eldadshulman
Session Name: endVsStart_chr1_heart
Genome Assembly: mm10
Creation Date: 2018-05-03
Views: 1571
|
1525334400 |
1571 |
Screenshot not available
Click Here to view |
Description:
Author: eldadshulman
Session Name: apa_heart
Genome Assembly: mm10
Creation Date: 2018-05-03
Views: 1504
|
1525334400 |
1504 |
 |
Description:
Author: ruhollah
Session Name: hg19_148Chr2_nmt3
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1567
|
1547452800 |
1567 |
 |
Description:
Author: che7oz
Session Name: hg19-IRF8-DCCC
Genome Assembly: hg19
Creation Date: 2018-10-03
Views: 1823
|
1538553600 |
1823 |
 |
Description:
Author: estercastillo
Session Name: Reprogenetics_PGD
Genome Assembly: hg19
Creation Date: 2018-04-27
Views: 1761
|
1524816000 |
1761 |
 |
Description:
Author: lob5149
Session Name: TSC2
Genome Assembly: hg19
Creation Date: 2018-04-26
Views: 2000
|
1524729600 |
2000 |
 |
Description:
Author: lob5149
Session Name: AKT1
Genome Assembly: hg19
Creation Date: 2018-04-26
Views: 1778
|
1524729600 |
1778 |
 |
Description:
Author: lob5149
Session Name: MTOR
Genome Assembly: hg19
Creation Date: 2018-04-26
Views: 1725
|
1524729600 |
1725 |
 |
Description:
Author: estercastillo
Session Name: JuanMartinez_LifeSeq_AS
Genome Assembly: hg19
Creation Date: 2018-04-22
Views: 1950
|
1524384000 |
1950 |
 |
Description:
Author: prutten
Session Name: rs6702619_3C-Seq_2replicates
Genome Assembly: hg19
Creation Date: 2015-03-17
Views: 1868
|
1426579200 |
1868 |
 |
Description:
Author: estercastillo
Session Name: Lara_AS_LaPaz
Genome Assembly: hg19
Creation Date: 2018-04-19
Views: 1881
|
1524124800 |
1881 |
 |
Description:
Author: roseaux12
Session Name: jinliying_nk
Genome Assembly: hg19
Creation Date: 2018-08-22
Views: 1884
|
1534924800 |
1884 |
 |
Description:
Author: celiagonzalez98
Session Name: class 3 part 1
Genome Assembly: hg19
Creation Date: 2020-02-12
Views: 1405
|
1581494400 |
1405 |
 |
Description:
Author: arthurcheng
Session Name: hg38 ATAC-seq
Genome Assembly: hg38
Creation Date: 2019-01-23
Views: 1071
|
1548230400 |
1071 |
 |
Description:
Author: riguang zhao
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2019-03-12
Views: 1608
|
1552377600 |
1608 |
 |
Description:
Author: Xavier Rambout
Session Name: mm9 2019-03-11
Genome Assembly: mm9
Creation Date: 2019-03-11
Views: 1449
|
1552291200 |
1449 |
 |
Description:
Author: ruhollah
Session Name: hg19_128Chr19+167Chr19_nmt11+11
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1616
|
1547452800 |
1616 |
 |
Description:
Author: Camila
Session Name: Enhancers-BreastCancersCells
Genome Assembly: hg19
Creation Date: 2018-04-21
Views: 1733
|
1524297600 |
1733 |
 |
Description:
Author: micca
Session Name: mm9_Buecker2014
Genome Assembly: mm9
Creation Date: 2018-04-03
Views: 1741
|
1522742400 |
1741 |
 |
Description: This session shows the ALDH2 gene, one of the two genes associated with alcohol intolerance in East Asian populations, in its entirety. Only the "UCSC Genes" track is enabled, showing two of the gene's potential isoforms.
Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: hg19_ALDH2
Genome Assembly: hg19
Creation Date: 2022-07-21
Views: 784
|
1658390400 |
784 |
 |
Description:
Author: Jcotney
Session Name: Mouse_chromhmm_15_18_state
Genome Assembly: mm10
Creation Date: 2018-10-20
Views: 852
|
1540022400 |
852 |
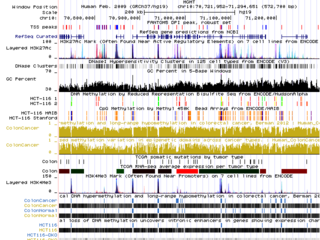 |
Description:
Author: arem
Session Name: colon meth
Genome Assembly: hg19
Creation Date: 2019-03-06
Views: 1528
|
1551859200 |
1528 |
 |
Description:
Author: jtm
Session Name: shiny_app_hg38_v0
Genome Assembly: hg38
Creation Date: 2018-03-28
Views: 4326
|
1522224000 |
4326 |
 |
Description:
Author: Hanson258
Session Name: DNA Damage ChIP-seq
Genome Assembly: hg38
Creation Date: 2018-07-17
Views: 2430
|
1531814400 |
2430 |
 |
Description:
Author: ldistefano
Session Name: dm6_DiStefanoData
Genome Assembly: dm6
Creation Date: 2019-03-12
Views: 1516
|
1552377600 |
1516 |
Screenshot not available
Click Here to view |
Description:
Author: jtm
Session Name: shiny_app_mm10_v0
Genome Assembly: mm10
Creation Date: 2018-04-04
Views: 2320
|
1522828800 |
2320 |
 |
Description:
Author: cocopow
Session Name: hg38_nucs
Genome Assembly: hg38
Creation Date: 2019-04-23
Views: 1547
|
1556006400 |
1547 |
 |
Description: The genes HBB and HBD are both expressed in red blood cells, but HBB is also expressed in many other tissues, whereas HBD is expressed in red cells almost exclusively.
Author: videoDemo1
Session Name: hg19_hemoglobin
Genome Assembly: hg19
Creation Date: 2018-03-13
Views: 1825
|
1520928000 |
1825 |
 |
Description:
Author: nehoralevi
Session Name: mm10-CTCF
Genome Assembly: mm10
Creation Date: 2019-06-03
Views: 2374
|
1559548800 |
2374 |
 |
Description: The GeneHancer database links human regulatory elements (enhancers and promoters) as arcs to their inferred target genes. Over 1 million regulatory elements obtained from seven genome-wide databases by GeneHancer are visualizable on the human hg19 and hg38 assemblies as color-coded curves ending on their respective targets. Read more about GeneHancer data on the track description page and this article <a href="https://www.ncbi.nlm.nih.gov/pubmed/28605766">GeneHancer: genome-wide integration of enhancers and target genes in GeneCards.</a>
Author: PublicSessions
Session Name: GeneHancer
Genome Assembly: hg38
Creation Date: 2019-05-31
Views: 1938
|
1559289600 |
1938 |
 |
Description:
Author: rafal.czapiewski@ed.ac.uk
Session Name: extended BACs and FOSMIDs
Genome Assembly: mm10
Creation Date: 2019-03-27
Views: 1846
|
1553673600 |
1846 |
 |
Description:
Author: AnaïsTest
Session Name: test
Genome Assembly: hg38
Creation Date: 2019-06-06
Views: 2032
|
1559808000 |
2032 |
 |
Description:
Author: brianlee
Session Name: test
Genome Assembly: hg38
Creation Date: 2019-06-06
Views: 1851
|
1559808000 |
1851 |
 |
Description:
Author: emmawentworth
Session Name: hg19-PWS
Genome Assembly: hg19
Creation Date: 2019-06-05
Views: 1925
|
1559721600 |
1925 |
 |
Description:
Author: cocopow
Session Name: mm10_altStrains
Genome Assembly: mm10
Creation Date: 2019-06-05
Views: 1555
|
1559721600 |
1555 |
 |
Description:
Author: evan_pepper
Session Name: extra_tRNAs_Analysis
Genome Assembly: hg19
Creation Date: 2018-03-08
Views: 1874
|
1520496000 |
1874 |
 |
Description:
Author: hchen17
Session Name: 20180307_1300_RESULTS_6pelvicFinStickleOut
Genome Assembly: hub_207981_oryLat03
Creation Date: 2018-03-07
Views: 1738
|
1520409600 |
1738 |
 |
Description:
Author: lboteva
Session Name: mm10_rif1_HP_
Genome Assembly: mm10
Creation Date: 2019-05-30
Views: 1794
|
1559203200 |
1794 |
 |
Description:
Author: lbennett
Session Name: impdh1retina
Genome Assembly: hg19
Creation Date: 2018-03-06
Views: 1811
|
1520323200 |
1811 |
 |
Description:
Author: ruhollah
Session Name: 12_ruh
Genome Assembly: hg38
Creation Date: 2018-11-13
Views: 1645
|
1542096000 |
1645 |
 |
Description:
Author: ruhollah
Session Name: hg19_2_chr8
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1674
|
1543219200 |
1674 |
 |
Description:
Author: yezheng
Session Name: HiC_ChIP-seq_short
Genome Assembly: hg19
Creation Date: 2018-03-04
Views: 1870
|
1520150400 |
1870 |
 |
Description:
Author: holdenl
Session Name: danRer11 zebrafish CNV across strains
Genome Assembly: danRer11
Creation Date: 2018-03-03
Views: 2667
|
1520064000 |
2667 |
 |
Description:
Author: estercastillo
Session Name: MartaRodriguez_Reus
Genome Assembly: hg19
Creation Date: 2018-02-28
Views: 1983
|
1519804800 |
1983 |
 |
Description:
Author: BenjaminMartin02
Session Name: Stefanska_data
Genome Assembly: hg19
Creation Date: 2018-02-14
Views: 2174
|
1518595200 |
2174 |
 |
Description:
Author: labrozam
Session Name: CGEMS Yeast AL
Genome Assembly: sacCer3
Creation Date: 2016-11-05
Views: 2062
|
1478332800 |
2062 |
 |
Description:
Author: labrozam
Session Name: CGEMS Worm AL test
Genome Assembly: ce11
Creation Date: 2016-11-20
Views: 1917
|
1479628800 |
1917 |
 |
Description:
Author: labrozam
Session Name: CGEMS Mouse AL test
Genome Assembly: mm9
Creation Date: 2016-11-29
Views: 2587
|
1480406400 |
2587 |
 |
Description:
Author: labrozam
Session Name: CGEMS Human AL test
Genome Assembly: hg38
Creation Date: 2016-11-21
Views: 1930
|
1479715200 |
1930 |
 |
Description:
Author: labrozam
Session Name: Anna-rs72802342
Genome Assembly: hg19
Creation Date: 2017-10-31
Views: 2194
|
1509436800 |
2194 |
 |
Description:
Author: jwjiao
Session Name: H2ZKO_E16_WT_E16
Genome Assembly: hg38
Creation Date: 2018-02-26
Views: 1822
|
1519632000 |
1822 |
 |
Description:
Author: troy.dumenil@qimr.edu.au
Session Name: MMR_branchpoints-230218
Genome Assembly: hg19
Creation Date: 2018-02-22
Views: 1776
|
1519286400 |
1776 |
 |
Description:
Author: yierjin_2008
Session Name: KMS12BM
Genome Assembly: hg19
Creation Date: 2018-02-22
Views: 1866
|
1519286400 |
1866 |
 |
Description: This session is an example of using the Short Match track to search a sequence in the current view with the IUPAC codes. The input of WACMKGTW catches two matches.
The session also using a feature of the Base Position track. You can highlight specific nucleotides with that track and AAC are selected to be highlighted (note, there is also a step taken to check the box to show reverse complements of motifs).
This region you will see AACATGTT and another reverse strand AACCTGTA, which appears to work with the IUPAC codes: http://genome.ucsc.edu/goldenPath/help/iupac.html
The Base Track and the Short Match track allow you to investigate the sequence in the current view when zoomed in with these highlight and search features.
Author: brianlee
Session Name: hg38.WACMKGTW
Genome Assembly: hg38
Creation Date: 2018-02-21
Views: 1021
|
1519200000 |
1021 |
 |
Description:
Author: simonwang612
Session Name: worm-chip-L4
Genome Assembly: ce10
Creation Date: 2018-02-20
Views: 1848
|
1519113600 |
1848 |
 |
Description:
Author: simonwang612
Session Name: worm-chip
Genome Assembly: ce10
Creation Date: 2018-02-19
Views: 1938
|
1519027200 |
1938 |
 |
Description:
Author: estercastillo
Session Name: JudithArmstrong_predisposicio
Genome Assembly: hg19
Creation Date: 2018-02-18
Views: 1770
|
1518940800 |
1770 |
 |
Description:
Author: Mannu Walia
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2018-12-13
Views: 1775
|
1544688000 |
1775 |
 |
Description:
Author: AnUnroyalKing
Session Name: Gene_File_King
Genome Assembly: hg19
Creation Date: 2018-02-15
Views: 1747
|
1518681600 |
1747 |
 |
Description:
Author: cmhct7
Session Name: Mouse Quad Data
Genome Assembly: mm10
Creation Date: 2016-09-12
Views: 1982
|
1473667200 |
1982 |
 |
Description:
Author: clauhag
Session Name: ContactSites_mixedpopulations
Genome Assembly: hg19
Creation Date: 2016-12-18
Views: 2180
|
1482048000 |
2180 |
 |
Description:
Author: clauhag
Session Name: ContactSites_clones
Genome Assembly: hg19
Creation Date: 2016-12-18
Views: 2089
|
1482048000 |
2089 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr11
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1891
|
1532505600 |
1891 |
 |
Description:
Author: zunpengliu66
Session Name: WT and DGCR8_dex2 H3K9me3 ChIP-seq
Genome Assembly: hg19
Creation Date: 2018-04-11
Views: 1735
|
1523433600 |
1735 |
 |
Description:
Author: samross
Session Name: danRer7_48hpf_6dpfbrain_RRBS
Genome Assembly: danRer7
Creation Date: 2018-02-14
Views: 1756
|
1518595200 |
1756 |
 |
Description:
Author: Lorena8a
Session Name: trabajo genomics HSC
Genome Assembly: hg19
Creation Date: 2018-02-14
Views: 1879
|
1518595200 |
1879 |
 |
Description:
Author: oac7
Session Name: oac7_hg19_all_snvs
Genome Assembly: hg19
Creation Date: 2018-02-13
Views: 1892
|
1518508800 |
1892 |
 |
Description:
Author: AnUnroyalKing
Session Name: Ian_King_Settings
Genome Assembly: hg19
Creation Date: 2018-02-13
Views: 2022
|
1518508800 |
2022 |
 |
Description:
Author: marzoldr
Session Name: hg38 MAPT density graphs and such
Genome Assembly: hg38
Creation Date: 2018-02-13
Views: 1781
|
1518508800 |
1781 |
 |
Description:
Author: marzoldr
Session Name: hg38 MAPT alleles
Genome Assembly: hg38
Creation Date: 2018-02-13
Views: 1764
|
1518508800 |
1764 |
 |
Description:
Author: marzoldr
Session Name: hg19 RHO custom 1
Genome Assembly: hg19
Creation Date: 2018-02-13
Views: 1742
|
1518508800 |
1742 |
 |
Description: This assembly hub for the CG2 clonal zebrafish is part of a much larger collection of assembly hubs. The hub details can be found on the gateway page for this hub <a href="http://genome.ucsc.edu/cgi-bin/hgGateway?hubUrl=http://genome-test.cse.ucsc.edu/gbdb/hubs/genbank/vertebrate_other/hub.ncbi.txt&genome=GCA_001483285.1_CG2v1.0&position=lastDbPos" >found here.</a> The hub is Biosample: SAMN03964926 and has Assembly accession ID: GCA_001483285.1. <br>
<br>
The hub is is part of this much larger assembly hub collection, with a launching page <a href="http://genome-test.cse.ucsc.edu/~hiram/hubs/genbank/vertebrate_mammalian/vertebrate_mammalian.ncbi.html" >found here</a> (note these will launch on our test development site as this hub is considered a prototype and hasn't passed quality control). Once connected to the above hub you have access to all of these hubs, the launching page only makes it more easy to directly link to the Track Browsing page. When connected one can also go to the Gateway page (earlier link) and see a <b>Download files for this assembly hub:</b> section with a <code>wget</code> command to access all the files for these assembly hubs.
Author: brianlee
Session Name: CG2zebrafish
Genome Assembly: hub_89563_GCA_001483285.1_CG2v1.0
Creation Date: 2018-07-06
Views: 1772
|
1530864000 |
1772 |
 |
Description:
Author: AnUnroyalKing
Session Name: King_setting
Genome Assembly: hg19
Creation Date: 2018-02-15
Views: 2067
|
1518681600 |
2067 |
 |
Description:
Author: estercastillo
Session Name: pseudogens
Genome Assembly: hg19
Creation Date: 2018-01-31
Views: 2632
|
1517385600 |
2632 |
 |
Description:
Author: marylaws
Session Name: foxm1 chip comparison
Genome Assembly: hg19
Creation Date: 2018-02-07
Views: 1749
|
1517990400 |
1749 |
 |
Description:
Author: marylaws
Session Name: FOXM 231 and mcf7
Genome Assembly: hg18
Creation Date: 2018-02-07
Views: 2097
|
1517990400 |
2097 |
 |
Description: This session illustrates the meaning of columns in the axt file definition here, http://genome.ucsc.edu/goldenPath/help/axt.html
You can see that indeed if you subtract to find the differences in the primary organism coordinates (panTro4: 78679-77637 = 1,042) it will be different than the values in the aligning organism coordinates (hg19: 88869-87772 = 1,097), but in the primary assembly sequence line you can see where there isn't a match in more than one place (most notable gct---------------------------------------------------ctt) as seen in this alignment of hg19 to panTro4. See the mailing list question here: <a href="https://groups.google.com/a/soe.ucsc.edu/d/msg/genome/-ohoSCyPIT0/c5nP7NzOBQAJ">link</a>.
Author: brianlee
Session Name: hg19.panTro4
Genome Assembly: hg19
Creation Date: 2018-02-02
Views: 1722
|
1517558400 |
1722 |
 |
Description:
Author: marzoldr
Session Name: TAS2R38 (T1)
Genome Assembly: hg19
Creation Date: 2018-01-23
Views: 2024
|
1516694400 |
2024 |
 |
Description:
Author: yezheng
Session Name: ATAC-seq_ALA_mm9
Genome Assembly: mm9
Creation Date: 2018-01-25
Views: 1887
|
1516867200 |
1887 |
 |
Description:
Author: ascendgene
Session Name: hg19_chr16:21960001-22400001
Genome Assembly: hg19
Creation Date: 2018-01-21
Views: 1841
|
1516521600 |
1841 |
 |
Description:
Author: chouhj
Session Name: noCHX_2017
Genome Assembly: sacCer3
Creation Date: 2017-08-14
Views: 1856
|
1502697600 |
1856 |
 |
Description:
Author: chouhj
Session Name: 46Del_2016batch
Genome Assembly: sacCer3
Creation Date: 2017-01-05
Views: 1894
|
1483603200 |
1894 |
 |
Description:
Author: chouhj
Session Name: 11Del_2015batch
Genome Assembly: sacCer3
Creation Date: 2017-01-05
Views: 2093
|
1483603200 |
2093 |
 |
Description:
Author: chmalee
Session Name: hg38notableChrYFeatures
Genome Assembly: hg38
Creation Date: 2017-03-06
Views: 2066
|
1488787200 |
2066 |
 |
Description:
Author: ascendgene
Session Name: hg19_chr15:32,020,001-32,420,001
Genome Assembly: hg19
Creation Date: 2018-01-21
Views: 2364
|
1516521600 |
2364 |
 |
Description:
Author: FrankaRang
Session Name: FR180123_mm10_groupmeeting
Genome Assembly: mm10
Creation Date: 2018-01-23
Views: 2150
|
1516694400 |
2150 |
 |
Description:
Author: zheyangshan
Session Name: EGFR BOTH
Genome Assembly: hg19
Creation Date: 2018-12-14
Views: 1543
|
1544774400 |
1543 |
 |
Description:
Author: rnaguru
Session Name: mm9-pY-MEME
Genome Assembly: mm9
Creation Date: 2018-01-18
Views: 2003
|
1516262400 |
2003 |
 |
Description: This assembly hub has a sequence that translates into a fun message when looking at codon translations.
Author: brianlee
Session Name: PAG_FUN
Genome Assembly: hub_211929_yourGenome
Creation Date: 2018-01-16
Views: 1951
|
1516089600 |
1951 |
 |
Description:
Author: tommi.andreani
Session Name: Neil.Methylome.Coverage10.WT.Clone5.DKO.clone7
Genome Assembly: mm10
Creation Date: 2018-01-12
Views: 1756
|
1515744000 |
1756 |
 |
Description:
Author: marzoldr
Session Name: Spring18 Yeast Test DMLJ
Genome Assembly: sacCer3
Creation Date: 2018-01-09
Views: 1734
|
1515484800 |
1734 |
 |
Description: This session shows TRPV3 which researchers believe is involved in our ability to sense and detect warm temperature. Scripps Research Institute scientists have demonstrated that mice lacking the TRPV3 protein have specific deficiencies in their ability to detect temperatures.
Author: brianlee
Session Name: skin_heat_sense
Genome Assembly: hg19
Creation Date: 2022-01-03
Views: 685
|
1641196800 |
685 |
 |
Description:
Author: megas
Session Name: hg19 Chip-Seq Encode vs mm10
Genome Assembly: hg19
Creation Date: 2018-01-02
Views: 1945
|
1514880000 |
1945 |
 |
Description: test the details .
Author: greece2019
Session Name: hg19_b0b1
Genome Assembly: hg19
Creation Date: 2019-03-03
Views: 1791
|
1551600000 |
1791 |
 |
Description:
Author: jkrakowiak
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2017-12-18
Views: 2317
|
1513584000 |
2317 |
 |
Description:
Author: jwjiao
Session Name: rbm3
Genome Assembly: mm10
Creation Date: 2017-12-17
Views: 1774
|
1513497600 |
1774 |
 |
Description: CircRNA and parental gene annotation as identified in the publication "Evolutionarily young transposable elements drive circular RNA repertoires" by Gruhl et al. (in preparation). The browser view include an additional track with the repeat dimers surrounding the respective circRNA.
Author: Frenzchen
Session Name: rheMac2 circRNA annotation
Genome Assembly: rheMac2
Creation Date: 2016-09-25
Views: 1343
|
1474790400 |
1343 |
 |
Description:
Author: mazawm
Session Name: hg#19RHO
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1547
|
1542096000 |
1547 |
 |
Description:
Author: Heywhoyou22
Session Name: Nrl and wt Cbr sites
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1611
|
1542096000 |
1611 |
 |
Description:
Author: nedbrewer66
Session Name: NAT7
Genome Assembly: hg38
Creation Date: 2018-12-28
Views: 1611
|
1545984000 |
1611 |
 |
Description:
Author: caojun
Session Name: Amr-20161031
Genome Assembly: mm9
Creation Date: 2016-10-31
Views: 1998
|
1477900800 |
1998 |
 |
Description:
Author: butlertj3
Session Name: nuc_g4
Genome Assembly: hg38
Creation Date: 2018-06-05
Views: 2001
|
1528185600 |
2001 |
 |
Description:
Author: butlertj3
Session Name: mtDNA_G4_homo_hetero
Genome Assembly: hg38
Creation Date: 2018-04-10
Views: 1910
|
1523347200 |
1910 |
 |
Description:
Author: brianlee
Session Name: sacCer3_spots
Genome Assembly: sacCer3
Creation Date: 2017-10-31
Views: 1964
|
1509436800 |
1964 |
 |
Description:
Author: brianlee
Session Name: hg38_interact_example
Genome Assembly: hg38
Creation Date: 2018-11-05
Views: 1669
|
1541404800 |
1669 |
 |
Description:
Author: brianlee
Session Name: hg38_GTEX_SUN
Genome Assembly: hg38
Creation Date: 2018-04-13
Views: 1834
|
1523606400 |
1834 |
 |
Description:
Author: brianlee
Session Name: hg38.exampleTracks
Genome Assembly: hg38
Creation Date: 2017-06-20
Views: 1811
|
1497945600 |
1811 |
 |
Description:
Author: brianlee
Session Name: hg38.SIRT1.relatedExpression
Genome Assembly: hg38
Creation Date: 2017-05-23
Views: 2293
|
1495526400 |
2293 |
 |
Description:
Author: brianlee
Session Name: hg19.pgSnpExample
Genome Assembly: hg19
Creation Date: 2017-07-19
Views: 1781
|
1500451200 |
1781 |
 |
Description:
Author: brianlee
Session Name: hg19.longTabix
Genome Assembly: hg19
Creation Date: 2017-05-24
Views: 1878
|
1495612800 |
1878 |
 |
Description: This session shows a search of primers. A researcher had an issue building PCR primers and Browser staff discovered that a desired reverse primer falls into an area of repeats. The original primer at 20 bases was near the limit of BLAT's sensitivity. With such short sequences, if BLAT has an over-used tile, a section of the query used to match and trigger an alignment, the algorithm will not be able to seed and extend an original BLAT hit in an area of repeats, which was what was likely happening to the reverse primer sequence in this session. In the session a larger BLAT query will result in a hit. Since primers are typically chosen from unique locations, Browser staff suggested it might be best to avoid a repeat-region, and noted that the nearby chr13:50,593,214-50,593,267 section appears to not have any repeats.
Author: brianlee
Session Name: Primers Blat Search
Genome Assembly: hg19
Creation Date: 2013-07-23
Views: 599
|
1374566400 |
599 |
 |
Description: This FORWARD strand session demonstrates how a user was using UCSC Genome tables to obtain exon sequences and their annotation and encountered a number of perceived discrepancies between the sequence data and the annotation, specifically asking "Why is the start position in the annotation consistently 1 nucleotide position before the start position for the sequence?" The response is that exon range provided with the sequence is 1-based because it is in position format, (chr#:##-##). While getting output as "all fields..." you see BED (chr# ## ##), 0-based format. This session shows how using the "all fields" output, we see this region full exon has exonStarts: 175629079, (but cdsEnd 175629122) with corresponding exonEnds:175629181. This is a reverse strand gene, so the last exon coordinates are really the beginning of the gene's first exon. So the applicable exonFrame items are the last two items "...,1,0,". But the last 0 here applies to the beginning of coding, cdsEnd coordinates (actually coding start since this is reverse strand). The second to last exonFrame, 1, refers to the exonStarts coordinates, 175629079, for an alanine, indicating exon2 picks up one nucleotide from the end of exon1.
Author: brianlee
Session Name: Example Session exonStarts exonEnds FORWARD
Genome Assembly: hg19
Creation Date: 2013-08-01
Views: 496
|
1375344000 |
496 |
 |
Description: This REVERSE strand session demonstrates how a user was using UCSC Genome tables to obtain exon sequences and their annotation and encountered a number of perceived discrepancies between the sequence data and the annotation, specifically asking "Why is the start position in the annotation consistently 1 nucleotide position before the start position for the sequence?" The response is that exon range provided with the sequence is 1-based because it is in position format, (chr#:##-##). While getting output as "all fields..." you see BED (chr# ## ##), 0-based format. This session shows how using the "all fields" output, we see this region full exon has exonStarts: 175629079, (but cdsEnd 175629122) with corresponding exonEnds:175629181. This is a reverse strand gene, so the last exon coordinates are really the beginning of the gene's first exon. So the applicable exonFrame items are the last two items "...,1,0,". But the last 0 here applies to the beginning of coding, cdsEnd coordinates (actually coding start since this is reverse strand). The second to last exonFrame, 1, refers to the exonStarts coordinates, 175629079, for an alanine, indicating exon2 picks up one nucleotide from the end of exon1.
Author: brianlee
Session Name: Example Session exonStarts exonEnds
Genome Assembly: hg19
Creation Date: 2013-07-31
Views: 436
|
1375257600 |
436 |
 |
Description:
Author: bisrat
Session Name: mm9
Genome Assembly: mm9
Creation Date: 2017-12-14
Views: 1835
|
1513238400 |
1835 |
 |
Description:
Author: bisrat
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2018-09-18
Views: 2092
|
1537257600 |
2092 |
 |
Description:
Author: benbeijs
Session Name: MYBPC1 (JSB) CM Data
Genome Assembly: hg19
Creation Date: 2018-09-04
Views: 1684
|
1536048000 |
1684 |
 |
Description:
Author: benbeijs
Session Name: ENCODE Exercise, 3
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1642
|
1542096000 |
1642 |
 |
Description:
Author: benbeijs
Session Name: ENCODE Exercise, 2
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1659
|
1542096000 |
1659 |
 |
Description:
Author: benbeijs
Session Name: ENCODE Exercise
Genome Assembly: hg19
Creation Date: 2018-11-13
Views: 1646
|
1542096000 |
1646 |
 |
Description:
Author: benbeijs
Session Name: DSP (JSB) CM Data
Genome Assembly: hg19
Creation Date: 2018-09-05
Views: 1711
|
1536134400 |
1711 |
 |
Description:
Author: benbeijs
Session Name: AMD SNP (JSB), Exercise 3
Genome Assembly: hg19
Creation Date: 2018-10-02
Views: 1621
|
1538467200 |
1621 |
 |
Description:
Author: tommi.andreani
Session Name: Neil.Methylome.Coverage.Higher.10
Genome Assembly: mm10
Creation Date: 2017-12-12
Views: 1822
|
1513065600 |
1822 |
 |
Description:
Author: nicklewis
Session Name: AMD SNP Second Try
Genome Assembly: hg19
Creation Date: 2017-11-02
Views: 1732
|
1509609600 |
1732 |
 |
Description:
Author: nicklewis
Session Name: AMD SNP Custom Tracks
Genome Assembly: hg38
Creation Date: 2017-10-31
Views: 1745
|
1509436800 |
1745 |
 |
Description:
Author: cocapo
Session Name: hg38_multiWig_noOverlay
Genome Assembly: hg38
Creation Date: 2018-12-13
Views: 1613
|
1544688000 |
1613 |
 |
Description:
Author: roseaux12
Session Name: 3_subset_nk
Genome Assembly: hg19
Creation Date: 2018-12-13
Views: 1681
|
1544688000 |
1681 |
 |
Description:
Author: abner_lim
Session Name: OVcar
Genome Assembly: hg19
Creation Date: 2018-12-09
Views: 1657
|
1544342400 |
1657 |
 |
Description:
Author: GenePeeks
Session Name: .2017.12 60bp Multi-Seq-Alg
Genome Assembly: hg19
Creation Date: 2017-12-10
Views: 1656
|
1512892800 |
1656 |
 |
Description:
Author: rnkeith
Session Name: dm6_first_ribo-seq
Genome Assembly: dm6
Creation Date: 2019-05-02
Views: 1540
|
1556784000 |
1540 |
 |
Description:
Author: cath
Session Name: v440hgw0Test
Genome Assembly: hg38
Creation Date: 2016-10-25
Views: 1861
|
1477382400 |
1861 |
 |
Description:
Author: cath
Session Name: MLQ19469
Genome Assembly: hg19
Creation Date: 2017-05-24
Views: 2179
|
1495612800 |
2179 |
 |
Description: These are the 12 UCSC Browser tracks for the RNA-seq experiment in which humanized and WT yeast were subjected to 1 hour of splicing inhibitors. There are six libraries in duplicate.
humanized hsh155 with DMSO carrier
humanized hsh155 with 5 uM pladienolide-b
humanized hsh155 with 0.5 uM pladienolide-b
humanized hsh155 with 5 uM thailanstatin-a
WT Hsh155 with DMSO carrier
WT Hsh155 with 5 uM pladienolide-b
Author: ohunter
Session Name: Hunter_et_al_sacCer3_Yeast_Splicing_Inhibition
Genome Assembly: sacCer3
Creation Date: 2023-07-25
Views: 596
|
1690272000 |
596 |
 |
Description:
Author: cocopow
Session Name: hg18_44way
Genome Assembly: hg18
Creation Date: 2019-07-24
Views: 1621
|
1563955200 |
1621 |
 |
Description:
Author: kinerettaler
Session Name: danRer11
Genome Assembly: danRer11
Creation Date: 2018-12-30
Views: 1614
|
1546156800 |
1614 |
 |
Description: The GeneHancer track relates enhancer and promoters to their interactions with nearby genes. The interactions track in the pack setting allows for a pack view of the data with the direction and name of individual endpoints clearly displayed. A highlighted GH01J209814 enhancer is associated with the gene IRF6 (Interferon Regulatory Factor 6) located about 10kb upstream from the gene and harbors regulatory non-coding variants strongly associated to Van Der Woude Syndrome 1 (VWS1), a disease involving cleft lip and cleft palate.
Author: view
Session Name: GeneHancerPack
Genome Assembly: hg19
Creation Date: 2019-03-29
Views: 1478
|
1553846400 |
1478 |
 |
Description: The JASPAR 2018 TFB Public Hub represents genome-wide predicted binding sites for TF (transcription factor) binding profiles in the JASPAR CORE vertebrates collection. Please click into the Track Description pages to see more details and credits and contacts for the hub's data.
Author: brianlee
Session Name: hg38.JASPARhub
Genome Assembly: hg38
Creation Date: 2017-09-29
Views: 2474
|
1506672000 |
2474 |
 |
Description: With this session loaded navigate to the Variant Annotation Integrator (VAI) tool (under the top blue bar Tools menu). Then scroll down and click the "Get results" button to experience how the VAI tool can now output HGVS terms. This session is loaded with an artificial input of variants on the human hg38 assembly with NCBI RefSeq Genes track (curated NM_*, NR_*, and YP_* subset) filtered for coding exons and splice site roles and on DNase regulatory elements with output in HTML Variant Effect Predictor format where the "Extra" column will include HGVS notation. Please note that a semi-colon ";" will separate results in that field (HGVSG=NC_000010.11:g.27005592_27005594delAGA; HGVSCN=NM_014915.2:c.5129_5131delTCT; EXON=34/34).
Author: brianlee
Session Name: hg38.exampleVAI
Genome Assembly: hg38
Creation Date: 2017-09-25
Views: 4644
|
1506326400 |
4644 |
 |
Description:
Author: rasou2ba
Session Name: Bio 630 CRX variants Rasoul
Genome Assembly: hg38
Creation Date: 2017-09-27
Views: 1703
|
1506499200 |
1703 |
 |
Description:
Author: lianeslaughter
Session Name: mm9-Locus3_primerDesign
Genome Assembly: mm9
Creation Date: 2017-09-26
Views: 1810
|
1506412800 |
1810 |
 |
Description:
Author: Cong Guo
Session Name: IDEAS Blank
Genome Assembly: hg19
Creation Date: 2017-09-26
Views: 1670
|
1506412800 |
1670 |
 |
Description:
Author: Lou
Session Name: pisOchAsseblyHub
Genome Assembly: hub_1100339_12Jun2017_28pcJ
Creation Date: 2019-04-22
Views: 1802
|
1555920000 |
1802 |
 |
Description:
Author: peytonse
Session Name: Bio 630 CRX variants Peyton
Genome Assembly: hg38
Creation Date: 2017-09-20
Views: 1768
|
1505894400 |
1768 |
 |
Description:
Author: nickcochran
Session Name: BE2C_MAPT_9-18-17
Genome Assembly: hg19
Creation Date: 2017-09-18
Views: 2067
|
1505721600 |
2067 |
 |
Description:
Author: morotz
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2017-09-14
Views: 1857
|
1505376000 |
1857 |
 |
Description:
Author: EvansLab
Session Name: EY_ATAC_Ileum
Genome Assembly: mm9
Creation Date: 2017-09-12
Views: 1772
|
1505203200 |
1772 |
 |
Description:
Author: mcgaughey
Session Name: mm10_retina_epigenetics
Genome Assembly: mm10
Creation Date: 2017-08-11
Views: 2034
|
1502438400 |
2034 |
 |
Description:
Author: yezheng
Session Name: HiC_ChIP-seq
Genome Assembly: hg19
Creation Date: 2018-03-02
Views: 1751
|
1519977600 |
1751 |
 |
Description:
Author: leggeteg
Session Name: Leggett, BIO480Uba1 example
Genome Assembly: hg19
Creation Date: 2017-09-06
Views: 1770
|
1504684800 |
1770 |
 |
Description:
Author: ofery1984
Session Name: FINAL Meth + RNA
Genome Assembly: mm10
Creation Date: 2017-09-06
Views: 1154
|
1504684800 |
1154 |
 |
Description:
Author: mccarthyti
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2017-08-30
Views: 1954
|
1504080000 |
1954 |
 |
Description:
Author: jcarlevaro
Session Name: TE_paper_hg38
Genome Assembly: hg38
Creation Date: 2017-08-28
Views: 1743
|
1503907200 |
1743 |
 |
Description: We are pleased to add the UniBind Hub to our list of publicly available track hubs. UniBind is a comprehensive map of direct transcription factor - DNA interactions in the human genome. UniBind expands on the previous data and is now available on GRCh38/hg38. Below is a session highlighting UniBind transcription factor-DNA binding interactions in the SPR gene.
http://genome.ucsc.edu/s/dschmelt/UniBindSRF
More info on UniBind:
https://unibind.uio.no
We would like to thank the Mathelier Lab at the University of Oslo for creating this hub!
Author: dschmelt
Session Name: UniBindSRF
Genome Assembly: hg38
Creation Date: 2019-04-17
Views: 2509
|
1555488000 |
2509 |
 |
Description:
Author: oac7
Session Name: oac7_Problem_Set_2
Genome Assembly: hg19
Creation Date: 2018-03-01
Views: 1740
|
1519891200 |
1740 |
 |
Description: This data includes Bcl11b ChIP-seq result.
Author: liuxiaoqin
Session Name: bcl11b-liuxiaoqin
Genome Assembly: mm10
Creation Date: 2022-08-17
Views: 635
|
1660723200 |
635 |
 |
Description:
Author: aniclem
Session Name: Clem, BIO480 Uba 1 example
Genome Assembly: hg19
Creation Date: 2017-09-03
Views: 1829
|
1504425600 |
1829 |
 |
Description:
Author: EvansLab
Session Name: Eiji-ATACseq2
Genome Assembly: hg19
Creation Date: 2017-07-10
Views: 1963
|
1499673600 |
1963 |
 |
Description:
Author: EvansLab
Session Name: EY_ATAC_Intes_Organoids
Genome Assembly: mm9
Creation Date: 2017-07-26
Views: 2081
|
1501056000 |
2081 |
 |
Description: Visualization in UCSC Genome Browser of genome coverages of all generated next-generation sequencing data included in the manuscript ('A novel approach for DNA footprinting using short double-stranded cell-free DNA from plasma').
Author: jnmllr
Session Name: Short_cfDNA_seq_manuscript
Genome Assembly: hg19
Creation Date: 2024-02-13
Views: 327
|
1707811200 |
327 |
 |
Description:
Author: svadlamudi
Session Name: KSR2_7-24-17
Genome Assembly: hg19
Creation Date: 2017-07-24
Views: 1942
|
1500883200 |
1942 |
 |
Description:
Author: jwjiao
Session Name: H2AZ-ChIP-E13-Cortex
Genome Assembly: mm10
Creation Date: 2017-07-13
Views: 1972
|
1499932800 |
1972 |
 |
Description: This session is a response to question for research on Myc binding sites on the IDH1 gene promoter, with a desire to analyze the conservation of Myc binding sites between human and mouse.
In the session Transcription Factor ChIP-seq (161 factors) from ENCODE track is displayed and three MYC binding spots represented. This clustered transcription factor binding track can be sorted to display only certain factors like MYC. Below the MYC binding spots (wgEncodeRegTfbsClusteredV3 track) the conservation data from 100 assembly alignments are displayed (cons100way track), with the specific data from mouse selected. Further below that the Chain/Net data (placentalChainNet track) specific for mouse are also displayed. To see this question on our Mailing List click this link: https://groups.google.com/a/soe.ucsc.edu/d/msg/genome/94u1jx2bk6c/py3OD90WAQAJ
Author: brianlee
Session Name: hg19.mycBinding2
Genome Assembly: hg19
Creation Date: 2017-07-13
Views: 1845
|
1499932800 |
1845 |
 |
Description:
Author: cocopow
Session Name: rn6_23676
Genome Assembly: rn6
Creation Date: 2019-06-20
Views: 1728
|
1561017600 |
1728 |
 |
Description:
Author: cocopow
Session Name: hg19_hubs
Genome Assembly: hg19
Creation Date: 2019-06-18
Views: 1512
|
1560844800 |
1512 |
 |
Description:
Author: Adhil
Session Name: sacCer3_supercoiling
Genome Assembly: sacCer3
Creation Date: 2019-06-17
Views: 1558
|
1560758400 |
1558 |
 |
Description:
Author: Joey Riepsaame
Session Name: mm9 DNAseI digital Me1 Me3 Ac27 Pol2
Genome Assembly: mm9
Creation Date: 2015-07-14
Views: 1203
|
1436860800 |
1203 |
 |
Description:
Author: janzop
Session Name: hsa_all_cccDNA_HBx_mRNA
Genome Assembly: hg38
Creation Date: 2017-06-20
Views: 2508
|
1497945600 |
2508 |
 |
Description:
Author: phageghost
Session Name: glasslab_microglia_hg38
Genome Assembly: hg38
Creation Date: 2017-06-19
Views: 2004
|
1497859200 |
2004 |
 |
Description:
Author: phageghost
Session Name: glasslab_microglia_mm10
Genome Assembly: mm10
Creation Date: 2017-06-19
Views: 1758
|
1497859200 |
1758 |
 |
Description: This sessionView is a collection of tracks centered around gene expression. The display is organized with gene annotations first, followed by mRNA and smRNA evidence. Following these data are <a href="https://www.nature.com/articles/ng.2653" target="_blank">GTEx tracks</a> which display median gene expression and transcripts from 51 tissues and 2 cells lines. The final tracks in this collection provide additional expression support: TSS/Dnase/chromatin state. The region visualized is a ~10,200bp window covering two <a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1891803/" target="_blank">HOX genes</a> which are regulators for embryonic development and continue to be expressed throughout postnatal life.
Author: view
Session Name: Expression
Genome Assembly: hg19
Creation Date: 2018-10-08
Views: 1757
|
1538985600 |
1757 |
 |
Description:
Author: nickcochran
Session Name: BE2C-Candidates-6-9-17
Genome Assembly: hg19
Creation Date: 2017-06-09
Views: 1860
|
1496995200 |
1860 |
 |
Description:
Author: ezra abramms
Session Name: nebulin bam site two
Genome Assembly: mm10
Creation Date: 2017-06-05
Views: 1980
|
1496649600 |
1980 |
 |
Description:
Author: wenzelmk
Session Name: BIO 481 hg38 CHR2 Chimp, Mouse, Chicken, Zebrafish
Genome Assembly: hg38
Creation Date: 2017-10-10
Views: 1804
|
1507622400 |
1804 |
 |
Description:
Author: peijinlim
Session Name: mm9 Nrf2 ChIPseq Mrp2
Genome Assembly: mm9
Creation Date: 2017-06-01
Views: 1860
|
1496304000 |
1860 |
 |
Description:
Author: yang1221720
Session Name: KDM3A_splicing
Genome Assembly: hg38
Creation Date: 2018-11-22
Views: 1598
|
1542873600 |
1598 |
 |
Description:
Author: ifollon
Session Name: hg19_sangre2
Genome Assembly: hg19
Creation Date: 2017-05-30
Views: 1958
|
1496131200 |
1958 |
 |
Description:
Author: jwrows2014
Session Name: EPAS1 search 5272017
Genome Assembly: hg38
Creation Date: 2017-05-27
Views: 1781
|
1495872000 |
1781 |
 |
Description: This session uses a new format called type=longTabix, here is a mailing list describing this type: https://groups.google.com/a/soe.ucsc.edu/d/msg/genome/kE2pIZUvfnA/jfopk7pFAQAJ
This new type uses a custom track that points to a .gz file, and where that is located there is also a .gz.tbi file that provides the index. The tab-separated .gz file has contents like the following: chr19 44116910 44119168 chr21:47876840-47878656,2 31515 .
The .tbi file is built using the tabix software from: http://samtools.sourceforge.net/tabix.shtml
Author: brianlee
Session Name: hg19.other_regions
Genome Assembly: hg19
Creation Date: 2017-05-25
Views: 1883
|
1495699200 |
1883 |
 |
Description:
Author: Fadak Alali
Session Name: Fall19 Yeast test FA
Genome Assembly: sacCer3
Creation Date: 2019-08-31
Views: 1435
|
1567238400 |
1435 |
 |
Description:
Author: ephong0305
Session Name: GeM MOA HD GWAS.v9
Genome Assembly: hg19
Creation Date: 2019-10-04
Views: 1535
|
1570176000 |
1535 |
 |
Description:
Author: Bowen
Session Name: hg38_HBB_rs334
Genome Assembly: hg38
Creation Date: 2019-10-24
Views: 1629
|
1571904000 |
1629 |
 |
Description:
Author: anneabraham1
Session Name: MAPT mutant alleles Anne Abraham
Genome Assembly: hg38
Creation Date: 2018-02-12
Views: 1742
|
1518422400 |
1742 |
 |
Description:
Author: XiangChen
Session Name: Bio481 Chen AMD SNPs rs67538026
Genome Assembly: hg38
Creation Date: 2017-10-31
Views: 2361
|
1509436800 |
2361 |
 |
Description:
Author: heathhm
Session Name: FALL19 Yeast test HMH
Genome Assembly: sacCer3
Creation Date: 2019-08-30
Views: 1606
|
1567152000 |
1606 |
 |
Description:
Author: farshad
Session Name: BRCA_hg19_H3K27Ac_related_profiles20170517
Genome Assembly: hg19
Creation Date: 2017-05-17
Views: 2129
|
1495008000 |
2129 |
 |
Description:
Author: ekrobins
Session Name: TSS_CAGE
Genome Assembly: hg19
Creation Date: 2017-05-17
Views: 2486
|
1495008000 |
2486 |
 |
Description:
Author: nbosch
Session Name: CRIM1
Genome Assembly: hg19
Creation Date: 2017-05-15
Views: 1851
|
1494835200 |
1851 |
 |
Description:
Author: brianlee
Session Name: hg38.refSeq.UCSC
Genome Assembly: hg38
Creation Date: 2017-05-12
Views: 2019
|
1494576000 |
2019 |
 |
Description:
Author: Francesco_Ferrari
Session Name: rs7599312_SNP
Genome Assembly: hg19
Creation Date: 2017-10-17
Views: 1758
|
1508227200 |
1758 |
 |
Description:
Author: sajjeev
Session Name: progeria_JB
Genome Assembly: mm10
Creation Date: 2019-05-23
Views: 1987
|
1558598400 |
1987 |
 |
Description:
Author: suestring
Session Name: example_DCdiffCA
Genome Assembly: hub_432097_DC
Creation Date: 2019-05-22
Views: 1647
|
1558512000 |
1647 |
 |
Description:
Author: suestring
Session Name: hub_432097_ZebraFish
Genome Assembly: hub_432097_ZebraFish
Creation Date: 2019-05-28
Views: 2435
|
1559030400 |
2435 |
 |
Description: Main overview of the ARID1a gene. Find the story in UCSC's education module: https://genome.ucsc.edu/training/education
Author: education
Session Name: arid1a_home
Genome Assembly: hg19
Creation Date: 2022-08-19
Views: 925
|
1660896000 |
925 |
 |
Description:
Author: cocopow
Session Name: hg38_primates
Genome Assembly: hg38
Creation Date: 2019-05-09
Views: 1573
|
1557388800 |
1573 |
 |
Description:
Author: russiandash
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2019-05-13
Views: 4235
|
1557734400 |
4235 |
 |
Description:
Author: DearDanielxd
Session Name: mm10
Genome Assembly: mm10
Creation Date: 2017-04-25
Views: 1899
|
1493107200 |
1899 |
 |
Description:
Author: yayinano
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2017-04-21
Views: 2105
|
1492761600 |
2105 |
 |
Description:
Author: linzho
Session Name: Zoltan_SA3_chip_data
Genome Assembly: hg19
Creation Date: 2019-05-15
Views: 1604
|
1557907200 |
1604 |
 |
Description:
Author: swarnaseetha
Session Name: TET5hmcTriplicates
Genome Assembly: mm10
Creation Date: 2019-10-08
Views: 1509
|
1570521600 |
1509 |
 |
Description:
Author: i.bader@salk.at
Session Name: hg19_GJB2
Genome Assembly: hg19
Creation Date: 2019-05-15
Views: 1546
|
1557907200 |
1546 |
 |
Description:
Author: i.bader@salk.at
Session Name: hg19_SLC26A4
Genome Assembly: hg19
Creation Date: 2019-05-15
Views: 1584
|
1557907200 |
1584 |
 |
Description:
Author: sayloren
Session Name: hg19-chr12-probes
Genome Assembly: hg19
Creation Date: 2017-04-07
Views: 1999
|
1491552000 |
1999 |
 |
Description:
Author: GDJ_Duke
Session Name: SJC_BAC_Lib_GDJ_PER1_BAC
Genome Assembly: hg38
Creation Date: 2018-09-06
Views: 1754
|
1536220800 |
1754 |
 |
Description:
Author: liyq
Session Name: mm10_2016
Genome Assembly: mm10
Creation Date: 2017-03-18
Views: 1841
|
1489824000 |
1841 |
 |
Description:
Author: PublicSessions
Session Name: N-masked regions on the Y chromosome for hg38
Genome Assembly: hg38
Creation Date: 2017-03-14
Views: 2036
|
1489478400 |
2036 |
 |
Description:
Author: gartnerj928
Session Name: Devi_methyl_IP_tracks
Genome Assembly: mm10
Creation Date: 2017-03-13
Views: 2612
|
1489392000 |
2612 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_H3K9me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1956
|
1489132800 |
1956 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_H3K9ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 2231
|
1489132800 |
2231 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_H3K4me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1857
|
1489132800 |
1857 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_H3K4me2
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1816
|
1489132800 |
1816 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_H3K4me1
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1870
|
1489132800 |
1870 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_H3K36me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1842
|
1489132800 |
1842 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_H3K27me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1902
|
1489132800 |
1902 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_H3K27ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1887
|
1489132800 |
1887 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_H3K9me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 2050
|
1489132800 |
2050 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_H3K9ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1844
|
1489132800 |
1844 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_H3K4me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1830
|
1489132800 |
1830 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_H3K4me2
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1789
|
1489132800 |
1789 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_H3K4me1
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1827
|
1489132800 |
1827 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_H3K36me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1851
|
1489132800 |
1851 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_H3K27me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1890
|
1489132800 |
1890 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_H3K27ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1860
|
1489132800 |
1860 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_H3K9me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1801
|
1489132800 |
1801 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_H3K9ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1887
|
1489132800 |
1887 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_H3K4me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1799
|
1489132800 |
1799 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_H3K4me2
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1796
|
1489132800 |
1796 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_H3K4me1
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1985
|
1489132800 |
1985 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_H3K36me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1796
|
1489132800 |
1796 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_H3K27me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1832
|
1489132800 |
1832 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_H3K27ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1965
|
1489132800 |
1965 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_H3K9me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1830
|
1489132800 |
1830 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_H3K9ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1993
|
1489132800 |
1993 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_H3K4me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1856
|
1489132800 |
1856 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_H3K4me2
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1855
|
1489132800 |
1855 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_H3K4me1
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1788
|
1489132800 |
1788 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_H3K36me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1766
|
1489132800 |
1766 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_H3K27me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1826
|
1489132800 |
1826 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_H3K27ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1828
|
1489132800 |
1828 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_CTCF
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1874
|
1489132800 |
1874 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_H3K9me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1934
|
1489132800 |
1934 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_H3K9ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1777
|
1489132800 |
1777 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_H3K4me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 2065
|
1489132800 |
2065 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_H3K4me2
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1987
|
1489132800 |
1987 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_H3K4me1
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1918
|
1489132800 |
1918 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_H3K36me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1761
|
1489132800 |
1761 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_H3K27me3
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 2231
|
1489132800 |
2231 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_H3K27ac
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1938
|
1489132800 |
1938 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_CTCF
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 2031
|
1489132800 |
2031 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_ALL
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1861
|
1489132800 |
1861 |
 |
Description:
Author: xulab
Session Name: tps_E16_ptm_ALL
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1815
|
1489132800 |
1815 |
 |
Description:
Author: xulab
Session Name: tps_E15_ptm_ALL
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1823
|
1489132800 |
1823 |
 |
Description:
Author: xulab
Session Name: tps_E14_ptm_ALL
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 1820
|
1489132800 |
1820 |
 |
Description:
Author: xulab
Session Name: tps_ALL_ptm_ALL
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 2610
|
1489132800 |
2610 |
 |
Description:
Author: ccornwel
Session Name: cfac
Genome Assembly: canFam3
Creation Date: 2016-09-26
Views: 1917
|
1474876800 |
1917 |
 |
Description:
Author: cocopow
Session Name: hg19-17q12
Genome Assembly: hg19
Creation Date: 2019-01-03
Views: 1721
|
1546502400 |
1721 |
 |
Description:
Author: rtmag4
Session Name: HCT116_ATAC_SEQ_TBLAB
Genome Assembly: hg38
Creation Date: 2017-03-03
Views: 1957
|
1488528000 |
1957 |
 |
Description:
Author: xulab
Session Name: tps_P0_ptm_CTCF
Genome Assembly: mm10
Creation Date: 2017-03-10
Views: 2234
|
1489132800 |
2234 |
 |
Description: Comparison of 7 primate genomes to the 2nd human chromosome.
Author: l.g.altizer
Session Name: 7Primate_Genomes
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 253
|
1726560000 |
253 |
 |
Description: Synteny and Conservation between Chimp and Human
Author: ALLDEREP
Session Name: Synteny and Conservation
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 217
|
1726560000 |
217 |
 |
Description:
Author: adriennelovett
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2017-02-24
Views: 1889
|
1487923200 |
1889 |
 |
Description:
Author: estercastillo
Session Name: AnaBelen_USalamanca_TP53
Genome Assembly: hg19
Creation Date: 2018-05-02
Views: 1895
|
1525248000 |
1895 |
 |
Description:
Author: Francesco_Ferrari
Session Name: AMLiPSC_H3K27Ac_ChIPseq_final_H3K27Ac
Genome Assembly: hg19
Creation Date: 2017-02-17
Views: 1881
|
1487318400 |
1881 |
 |
Description:
Author: jberus
Session Name: Bio2010-JB
Genome Assembly: hg18
Creation Date: 2017-02-16
Views: 1848
|
1487232000 |
1848 |
 |
Description:
Author: SB
Session Name: hg38_ALK E20 Cas9
Genome Assembly: hg38
Creation Date: 2017-02-15
Views: 1983
|
1487145600 |
1983 |
 |
Description: We have recently released a file type called "longTabix" which supports inter & intra-chromosomal interactions by drawing an "arc" among paired regions in the UCSC Genome Browser. Note the first track at the top of the display, you can click on any of the "arcs" or interaction connections to see details such as ID and score.
Credit goes to Cath Tyner for the creation of this session.
Link to the original question:
https://groups.google.com/a/soe.ucsc.edu/d/msg/genome/kE2pIZUvfnA/jfopk7pFAQAJ
Author: PublicSessions
Session Name: Viewing inter- & intra-chromosomal interactions
Genome Assembly: hg19
Creation Date: 2017-02-13
Views: 1739
|
1486972800 |
1739 |
 |
Description:
Author: Francesco_Ferrari
Session Name: AMLiPSC_H3K27Ac_ChIPseq_CORRECTED
Genome Assembly: hg19
Creation Date: 2017-02-10
Views: 1805
|
1486713600 |
1805 |
 |
Description:
Author: Francesco_Ferrari
Session Name: AMLiPSC_H3K27Ac_ChIPseq_CORRECTED_autoscaled
Genome Assembly: hg19
Creation Date: 2017-02-13
Views: 1828
|
1486972800 |
1828 |
 |
Description:
Author: Francesco_Ferrari
Session Name: AMLiPSC_H3K27Ac_ChIPseq_days_5_20_35_40
Genome Assembly: hg19
Creation Date: 2017-02-01
Views: 1836
|
1485936000 |
1836 |
 |
Description:
Author: thomas.sparks
Session Name: 181227-atac1-encode_dnase
Genome Assembly: mm10
Creation Date: 2018-12-27
Views: 1649
|
1545897600 |
1649 |
 |
Description:
Author: xrdong10
Session Name: hg19_fdek
Genome Assembly: hg19
Creation Date: 2017-01-24
Views: 1912
|
1485244800 |
1912 |
 |
Description:
Author: rikrdo89
Session Name: rna-seq cardiac
Genome Assembly: mm9
Creation Date: 2017-01-24
Views: 1895
|
1485244800 |
1895 |
 |
Description:
Author: Francesco_Ferrari
Session Name: AMLiPSC_H3K27Ac_ChIPseq
Genome Assembly: hg19
Creation Date: 2017-01-23
Views: 1819
|
1485158400 |
1819 |
 |
Description:
Author: ryansamuel13
Session Name: TAS2R28 Gene
Genome Assembly: hg19
Creation Date: 2017-01-23
Views: 1867
|
1485158400 |
1867 |
 |
Description:
Author: Shockwing
Session Name: South, Spring17, Uba1 example
Genome Assembly: mm9
Creation Date: 2017-01-16
Views: 1852
|
1484553600 |
1852 |
 |
Description:
Author: ryansamuel13
Session Name: Samuel, Spring17, Uba1 example
Genome Assembly: mm9
Creation Date: 2017-01-16
Views: 1778
|
1484553600 |
1778 |
 |
Description:
Author: anneabraham1
Session Name: Spring18 Yeast test ALA
Genome Assembly: sacCer3
Creation Date: 2018-01-08
Views: 1727
|
1515398400 |
1727 |
 |
Description:
Author: harri5al
Session Name: Spring18 Yeast Test (AH)
Genome Assembly: sacCer3
Creation Date: 2018-01-08
Views: 1811
|
1515398400 |
1811 |
 |
Description:
Author: keitermd
Session Name: Spring17_Correct MOUSE TEST - MDK
Genome Assembly: mm9
Creation Date: 2017-01-09
Views: 2035
|
1483948800 |
2035 |
 |
Description:
Author: khan2sa
Session Name: Spring17 Mouse test (SK)
Genome Assembly: mm9
Creation Date: 2017-01-09
Views: 2096
|
1483948800 |
2096 |
 |
Description:
Author: ryansamuel13
Session Name: Spring17 Worm test (RMS)
Genome Assembly: ce11
Creation Date: 2017-01-09
Views: 1792
|
1483948800 |
1792 |
 |
Description:
Author: keitermd
Session Name: Spring17 MOUSE TEST - MDK
Genome Assembly: mm10
Creation Date: 2017-01-09
Views: 1805
|
1483948800 |
1805 |
 |
Description:
Author: lesevimx
Session Name: Spring17 Mouse test (ML)
Genome Assembly: mm9
Creation Date: 2017-01-09
Views: 1778
|
1483948800 |
1778 |
 |
Description:
Author: idror
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2017-01-03
Views: 2081
|
1483430400 |
2081 |
 |
Description:
Author: yuelab
Session Name: mm9_min
Genome Assembly: mm9
Creation Date: 2017-01-16
Views: 1898
|
1484553600 |
1898 |
 |
Description:
Author: warrenac
Session Name: GIT-mQTL-ASM
Genome Assembly: hg19
Creation Date: 2016-10-04
Views: 2280
|
1475568000 |
2280 |
 |
Description:
Author: jkearns94
Session Name: Fall 16 481 Final JK
Genome Assembly: mm9
Creation Date: 2016-12-12
Views: 1945
|
1481529600 |
1945 |
 |
Description:
Author: nightfury
Session Name: mm9-fam60a-de
Genome Assembly: mm9
Creation Date: 2016-12-11
Views: 1980
|
1481443200 |
1980 |
 |
Description: Since the reference genome assembly is a series of clone sequences from multiple individuals, and all individuals contain rare alleles, some of these rare alleles are included in reference assemblies. In this case, NM_001103170 (TGc), "c" mismatches the reference assemblies "A" because the reference sequence contains a substitution at this location. Please note that this example is on hg18, many (but not all) of these cases have been addressed in newer assemblies. AADACL3, in particular, does not have this substitution in GRCh38/hg38.
Credit goes to Christopher Villarreal for creation of this session.
Link to original question:
https://groups.google.com/a/soe.ucsc.edu/d/msg/genome/6TG1Pze9rUI/TLsKg7bNAwAJ
Author: PublicSessions
Session Name: hg18.AADACL3_mRNA_discrepancy_vs_reference
Genome Assembly: hg18
Creation Date: 2016-12-09
Views: 1879
|
1481270400 |
1879 |
 |
Description:
Author: yezheng
Session Name: ATAC-seq_mm9_WT-+MU-+
Genome Assembly: mm9
Creation Date: 2019-01-31
Views: 1561
|
1548921600 |
1561 |
 |
Description:
Author: labrozam
Session Name: Labrozzi, Fall 17, Uba1 example
Genome Assembly: mm9
Creation Date: 2017-10-01
Views: 1716
|
1506844800 |
1716 |
 |
Description:
Author: cocopow
Session Name: hg19_bigBed12
Genome Assembly: hg19
Creation Date: 2019-03-21
Views: 1557
|
1553155200 |
1557 |
 |
Description:
Author: suestring
Session Name: ???
Genome Assembly: hub_162375_Pleuco
Creation Date: 2019-03-08
Views: 1519
|
1552032000 |
1519 |
 |
Description:
Author: suestring
Session Name: NotFusion
Genome Assembly: hub_162375_Pleuco
Creation Date: 2019-03-08
Views: 1619
|
1552032000 |
1619 |
 |
Description:
Author: suestring
Session Name: 2
Genome Assembly: hub_162375_Pleuco
Creation Date: 2019-03-08
Views: 1701
|
1552032000 |
1701 |
 |
Description:
Author: suestring
Session Name: 1
Genome Assembly: hub_162375_Pleuco
Creation Date: 2019-03-08
Views: 1651
|
1552032000 |
1651 |
 |
Description:
Author: arem
Session Name: hg19 fin
Genome Assembly: hg19
Creation Date: 2019-02-21
Views: 1874
|
1550736000 |
1874 |
 |
Description:
Author: sureshpsbio
Session Name: mm10_Chip fam40
Genome Assembly: mm10
Creation Date: 2019-02-20
Views: 2043
|
1550649600 |
2043 |
 |
Description:
Author: ruhollah
Session Name: hg19_1600_chr1_nmt=20
Genome Assembly: hg19
Creation Date: 2019-02-20
Views: 1534
|
1550649600 |
1534 |
 |
Description:
Author: ruhollah
Session Name: hg19_844_chr7_nmt20
Genome Assembly: hg19
Creation Date: 2019-02-20
Views: 1503
|
1550649600 |
1503 |
 |
Description:
Author: ruhollah
Session Name: hg19_412Chr8_nmt19
Genome Assembly: hg19
Creation Date: 2019-02-20
Views: 2104
|
1550649600 |
2104 |
 |
Description:
Author: ruhollah
Session Name: hg19_1148_chr1_nmt18
Genome Assembly: hg19
Creation Date: 2019-02-20
Views: 1571
|
1550649600 |
1571 |
 |
Description:
Author: liaohy
Session Name: RNAseq_Liu.DY_AM20181126-13
Genome Assembly: mm10
Creation Date: 2019-12-31
Views: 1441
|
1577779200 |
1441 |
 |
Description:
Author: ruhollah
Session Name: hg19_986_chr3_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1656
|
1554105600 |
1656 |
 |
Description:
Author: ruhollah
Session Name: hg19_1305_chr16_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1506
|
1554105600 |
1506 |
 |
Description:
Author: ruhollah
Session Name: hg19_1388_chr1_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1746
|
1554105600 |
1746 |
 |
Description:
Author: ruhollah
Session Name: hg19_1136_chr17_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1527
|
1554105600 |
1527 |
 |
Description:
Author: ruhollah
Session Name: hg19_1373_chr19_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1513
|
1554105600 |
1513 |
 |
Description:
Author: ruhollah
Session Name: hg19_1044_chr2_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1568
|
1554105600 |
1568 |
 |
Description:
Author: ruhollah
Session Name: hg19_128_chr14_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1519
|
1554105600 |
1519 |
 |
Description:
Author: ruhollah
Session Name: hg19_511_chr14_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1728
|
1554105600 |
1728 |
 |
Description:
Author: ruhollah
Session Name: hg19_1111_chr9_nmtf11
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1581
|
1554105600 |
1581 |
 |
Description:
Author: ruhollah
Session Name: hg19_237_chr22_nmtf12
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1785
|
1554105600 |
1785 |
 |
Description:
Author: ruhollah
Session Name: hg19_1327_chr17_nmtf12
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1735
|
1554105600 |
1735 |
 |
Description:
Author: ruhollah
Session Name: hg19_40_chr22_nmtf12
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1806
|
1554105600 |
1806 |
 |
Description:
Author: ruhollah
Session Name: hg19_954_chr1_nmtf12
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1563
|
1554105600 |
1563 |
 |
Description:
Author: ruhollah
Session Name: hg19_1129_chr17_nmtf12
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1709
|
1554105600 |
1709 |
 |
Description:
Author: ruhollah
Session Name: hg19_960_chrX_nmtf13
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1674
|
1554105600 |
1674 |
 |
Description:
Author: ruhollah
Session Name: hg19_662_chr6_nmtf13
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1494
|
1554105600 |
1494 |
 |
Description:
Author: ruhollah
Session Name: hg19_876_chr17_nmtf13
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1512
|
1554105600 |
1512 |
 |
Description:
Author: ruhollah
Session Name: hg19_149_chr1_nmtf13
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1532
|
1554105600 |
1532 |
 |
Description:
Author: bashamal
Session Name: basham hg38 assignment 10/10
Genome Assembly: hg38
Creation Date: 2017-10-09
Views: 1854
|
1507536000 |
1854 |
 |
Description:
Author: vperreau@unimelb.edu.au
Session Name: mm10 celltype NTRK2
Genome Assembly: mm10
Creation Date: 2018-12-04
Views: 924
|
1543910400 |
924 |
Screenshot not available
Click Here to view |
Description:
Author: Tao Zhang
Session Name: chicken transcriptome
Genome Assembly: hub_30425_galGal4
Creation Date: 2016-11-29
Views: 2494
|
1480406400 |
2494 |
 |
Description:
Author: chkcole
Session Name: TN5_Prime_Alignments
Genome Assembly: hg38
Creation Date: 2017-10-24
Views: 2324
|
1508832000 |
2324 |
Screenshot not available
Click Here to view |
Description:
Author: maria pia miano
Session Name: hg19s
Genome Assembly: hg19
Creation Date: 2016-11-16
Views: 2034
|
1479283200 |
2034 |
 |
Description:
Author: jtm
Session Name: shiny_app_mm10_v2
Genome Assembly: mm10
Creation Date: 2018-11-05
Views: 1927
|
1541404800 |
1927 |
 |
Description:
Author: headricn
Session Name: Fall16 481 final CH
Genome Assembly: mm9
Creation Date: 2016-12-11
Views: 1964
|
1481443200 |
1964 |
Screenshot not available
Click Here to view |
Description:
Author: Courtney Stout
Session Name: AMD hg38 APOE SNP (CS)
Genome Assembly: hg38
Creation Date: 2016-11-04
Views: 1939
|
1478246400 |
1939 |
Screenshot not available
Click Here to view |
Description:
Author: Courtney Stout
Session Name: hg38 AMD CNN2 SNP (CS)
Genome Assembly: hg38
Creation Date: 2016-11-04
Views: 1954
|
1478246400 |
1954 |
Screenshot not available
Click Here to view |
Description:
Author: Courtney Stout
Session Name: hg38 AMD APOE SNP(CS)
Genome Assembly: hg38
Creation Date: 2016-11-04
Views: 2027
|
1478246400 |
2027 |
Screenshot not available
Click Here to view |
Description:
Author: Courtney Stout
Session Name: hg38 AMD SNPs
Genome Assembly: hg38
Creation Date: 2016-10-27
Views: 1964
|
1477555200 |
1964 |
 |
Description: This session highlights the differences in microRNA expression between endothelial cells and other primary cells types. The data in this session comes from the "Towards the human cellular microRNAome" public hub, created by Marc Halushka and Arun Patil at the Johns Hopkins Medical Institute.
Author: chmalee
Session Name: hg38_microRNA_cell_expression
Genome Assembly: hg38
Creation Date: 2017-09-11
Views: 3129
|
1505116800 |
3129 |
 |
Description:
Author: troy.dumenil@qimr.edu.au
Session Name: MMR-branchpoints-260318
Genome Assembly: hg19
Creation Date: 2018-03-26
Views: 1813
|
1522051200 |
1813 |
 |
Description:
Author: rmaus
Session Name: nha_PE
Genome Assembly: mm8
Creation Date: 2016-10-18
Views: 1982
|
1476777600 |
1982 |
 |
Description:
Author: Sega666
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2016-10-19
Views: 1859
|
1476864000 |
1859 |
 |
Description:
Author: cath
Session Name: v339publicSessionTest
Genome Assembly: hg38
Creation Date: 2016-10-04
Views: 2306
|
1475568000 |
2306 |
Screenshot not available
Click Here to view |
Description:
Author: Isilver1
Session Name: PCBP2_CLIP-seq_Reps1-3
Genome Assembly: mm10
Creation Date: 2017-03-28
Views: 2123
|
1490688000 |
2123 |
 |
Description:
Author: Sega666
Session Name: 02.11
Genome Assembly: hg19
Creation Date: 2016-11-02
Views: 1945
|
1478073600 |
1945 |
 |
Description:
Author: artman1967
Session Name: hg19_ATAC_2019
Genome Assembly: hg19
Creation Date: 2019-03-28
Views: 1680
|
1553760000 |
1680 |
 |
Description:
Author: mirigitik
Session Name: bahcc1
Genome Assembly: mm9
Creation Date: 2016-05-16
Views: 2335
|
1463385600 |
2335 |
 |
Description:
Author: sergi.villatoro
Session Name: Pangaea Biotech
Genome Assembly: hg19
Creation Date: 2016-09-14
Views: 1973
|
1473840000 |
1973 |
 |
Description:
Author: zxtzhangqian
Session Name: 3a-tet2
Genome Assembly: mm9
Creation Date: 2016-09-14
Views: 1975
|
1473840000 |
1975 |
 |
Description:
Author: vivekgopalanmedgenome
Session Name: hg19_panel_design_JRK
Genome Assembly: hg19
Creation Date: 2016-09-11
Views: 1971
|
1473580800 |
1971 |
 |
Description:
Author: sergi.villatoro
Session Name: Pangaea Biotech GenneReader
Genome Assembly: hg19
Creation Date: 2016-09-16
Views: 1889
|
1474012800 |
1889 |
 |
Description:
Author: sushivision
Session Name: TKNK31-090116
Genome Assembly: mm9
Creation Date: 2016-09-02
Views: 1855
|
1472803200 |
1855 |
Screenshot not available
Click Here to view |
Description:
Author: lijunyao@usc.edu
Session Name: C2H2_type_1
Genome Assembly: hg19
Creation Date: 2016-09-01
Views: 1936
|
1472716800 |
1936 |
 |
Description:
Author: sambuca_shot
Session Name: mouse_reprog
Genome Assembly: mm10
Creation Date: 2016-03-23
Views: 2013
|
1458720000 |
2013 |
 |
Description: GTEx Allele Specific Expression hub from the Lappalainen Lab at the New York Genome Center. This session shows a region along chromosome 17 of high skin-specific allelic imbalance in a large number of Keratin genes.
Author: chmalee
Session Name: hg19KeratinAse
Genome Assembly: hg19
Creation Date: 2016-08-30
Views: 2968
|
1472544000 |
2968 |
 |
Description:
Author: pfiziev
Session Name: t_cell_development.H3K4me2 DECSTEP
Genome Assembly: mm9
Creation Date: 2016-08-26
Views: 2244
|
1472198400 |
2244 |
 |
Description:
Author: ps
Session Name: hg19.newTargets
Genome Assembly: hg19
Creation Date: 2016-08-26
Views: 2018
|
1472198400 |
2018 |
 |
Description: We sequenced the hermaphroditic freshwater snail, Biomphalaria glabrata (strain BB02), the host for the medically important trematode parasite, Schistosoma mansoni, using 1X coverage from plasmids and 0.1X BAC end sequencing on the ABI3730xl and 10X coverage sequenced on the Roche 454 Sequencer (including 2 paired end read runs) was initially assembled with Newbler. The Newbler assembly was screened for contamination then merged with a Soap assembly of Illumina reads followed by use of an in-house program for collapsing of redundant heterozygous contigs. Next, we applied our program, GapCloser, which closes gaps in the assembly by making iterative joins using Illumina reads. Finally we removed contigs less than 200 bases and incorporated reads into the assembly that had been assembled in a prior version of the Newbler assembly but were not assembled in this round. The resulting assembly, going by the name of assembly version 4.3, is 898.9Mb bases with an N50 contig length of 6.9kb and N50 scaffold length of 42kb. For creation of the linkage group AGP files, we identified all scaffolds that were uniquely placed on a single linkage group. Because of low marker density, only 145Mb was localized to specific linkage groups and scaffolds could not be ordered and oriented within linkage groups. Therefore, the scaffolds were simply placed in the order suggested by the linkage mapping on *_random for each linkage group.
Credits:
B. glabrata BB02 samples - Omar dos Santos Carvalho, Centro de Pesquisas Rene Rachou-Fiocruz (sample location: Barreiro, Brazil)
BAC library - Coen M. Adema et al. (2006)
Sequencing and Assembly - The Genome Institute, Washington University School of Medicine
Linkage map
- Jacob Tennessen and Michael Blouin, Oregon State University White Paper
- Matty Knight et al (2003) Others -
- Fred Lewis Biomedical Research Institute of the American Foundation of Biomedical Research
- Eric Loker University of New Mexico Biology
- Nithya Raghavan Biomedical Research Institute of the American Foundation of Biomedical Research
Author: lijing
Session Name: hub_107761_bioGla0
Genome Assembly: hub_107761_bioGla0
Creation Date: 2016-08-24
Views: 2072
|
1472025600 |
2072 |
 |
Description:
Author: elinshiao
Session Name: HEK293T_all
Genome Assembly: hg19
Creation Date: 2020-09-07
Views: 1758
|
1599465600 |
1758 |
 |
Description:
Author: mperelis
Session Name: Stein_PDX1
Genome Assembly: mm10
Creation Date: 2016-09-30
Views: 2097
|
1475222400 |
2097 |
 |
Description:
Author: treparriscos
Session Name: Sara_poised_enhancers_1xnorm_mm10_2
Genome Assembly: mm10
Creation Date: 2016-06-03
Views: 1487
|
1464940800 |
1487 |
 |
Description:
Author: sappingtonae
Session Name: test
Genome Assembly: hg38
Creation Date: 2016-08-15
Views: 2125
|
1471248000 |
2125 |
 |
Description:
Author: yezheng
Session Name: ATAC-seq_mm9
Genome Assembly: mm9
Creation Date: 2018-01-22
Views: 2275
|
1516608000 |
2275 |
 |
Description:
Author: robert-cornell@uiowa.edu
Session Name: Lidral, hg19, CL/P GWAS intervals
Genome Assembly: hg19
Creation Date: 2014-09-18
Views: 1780
|
1411027200 |
1780 |
 |
Description:
Author: shiguoming
Session Name: mm9_160812
Genome Assembly: mm9
Creation Date: 2016-08-11
Views: 1518
|
1470902400 |
1518 |
 |
Description:
Author: ykumar84
Session Name: mm10_Oct4
Genome Assembly: mm10
Creation Date: 2019-07-22
Views: 1752
|
1563782400 |
1752 |
 |
Description: This is an assembly hub for the AgamP4 assembly of Anopheles gambiae PEST strain. It includes the assembly; the coding genes and pseudogenes from vectorbase version 4.3; predicted stop codon readthrough regions; PhyloCSF tracks showing evolutionary protein-coding potential; splice-prediction tracks using the maximum-entropy splice-prediction algorithm; and novel coding and pseudogene predictions using PhyloCSF, excluding regions already annotated in vectorbase version 4.3.
Author: iljungr
Session Name: AgamP4
Genome Assembly: hub_102577_AgamP4
Creation Date: 2016-08-09
Views: 2580
|
1470729600 |
2580 |
 |
Description:
Author: brianlee
Session Name: mm9.RP23.470O14
Genome Assembly: mm9
Creation Date: 2016-08-03
Views: 1730
|
1470211200 |
1730 |
 |
Description:
Author: cocopow
Session Name: hub_1567195_araTha1_remap
Genome Assembly: hub_1567195_araTha1
Creation Date: 2019-09-04
Views: 1896
|
1567584000 |
1896 |
 |
Description:
Author: QAtester3
Session Name: ce11_ENCODE_hub
Genome Assembly: ce11
Creation Date: 2018-02-08
Views: 1666
|
1518076800 |
1666 |
 |
Description:
Author: QAtester3
Session Name: !@!$(GSDFG
Genome Assembly: galGal6
Creation Date: 2019-01-22
Views: 1697
|
1548144000 |
1697 |
 |
Description: This session of gene prediction tracks for "heart genes" uses the interact and bigGenePred formats to create decorations and the image of a heart. Read more about the interact format and bigGenePred formats from links on the Data File Formats help page under the Help menu and FAQs: http://genome.ucsc.edu/FAQ/FAQformat.html
Author: PublicSessions
Session Name: heart
Genome Assembly: hg38
Creation Date: 2019-02-11
Views: 4372
|
1549872000 |
4372 |
 |
Description:
Author: PublicSessions
Session Name: Tabula_muris
Genome Assembly: mm10
Creation Date: 2019-04-05
Views: 1694
|
1554451200 |
1694 |
 |
Description:
Author: cook2wj
Session Name: Alpha Syn Chip Data
Genome Assembly: hg18
Creation Date: 2016-07-22
Views: 1885
|
1469174400 |
1885 |
 |
Description:
Author: matthew.law
Session Name: MTAP_melanoma
Genome Assembly: hg19
Creation Date: 2016-07-25
Views: 2016
|
1469433600 |
2016 |
 |
Description:
Author: lesevimx
Session Name: Alpha Syn Chip Data
Genome Assembly: hg18
Creation Date: 2016-07-21
Views: 1939
|
1469088000 |
1939 |
 |
Description:
Author: jpate2
Session Name: CGEMS Yeast TRJP primers test
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 2203
|
1469088000 |
2203 |
 |
Description:
Author: hadani81
Session Name: CGEMS Mouse HD/MC/JD Test
Genome Assembly: mm9
Creation Date: 2016-07-21
Views: 1817
|
1469088000 |
1817 |
 |
Description:
Author: Nithesh
Session Name: CGEMS Yeast NPC test
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 1910
|
1469088000 |
1910 |
 |
Description:
Author: monroejd
Session Name: CGEMS Yeast JM
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 1850
|
1469088000 |
1850 |
 |
Description:
Author: jpate2
Session Name: CGEMS Yeast TRJP test
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 1877
|
1469088000 |
1877 |
 |
Description:
Author: kapsakcj
Session Name: CGEMS Mouse CK test
Genome Assembly: mm9
Creation Date: 2016-07-21
Views: 1818
|
1469088000 |
1818 |
 |
Description:
Author: Shockwing
Session Name: CGEMS Yeast (Shockwing) Test
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 2078
|
1469088000 |
2078 |
 |
Description:
Author: herrsp
Session Name: CGEMS Yeast SPH test
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 1871
|
1469088000 |
1871 |
 |
Description:
Author: blossta
Session Name: CGEMS Worm TB test
Genome Assembly: ce11
Creation Date: 2016-07-21
Views: 1913
|
1469088000 |
1913 |
 |
Description:
Author: shufengliu
Session Name: CGEMS Human SL test
Genome Assembly: hg38
Creation Date: 2016-07-21
Views: 2127
|
1469088000 |
2127 |
 |
Description:
Author: klj002
Session Name: CGEMS Worm (KLJ SFB) test
Genome Assembly: ce11
Creation Date: 2016-07-21
Views: 1863
|
1469088000 |
1863 |
 |
Description:
Author: MaxHenderson
Session Name: CGEMS Yeast MEH test
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 1915
|
1469088000 |
1915 |
 |
Description:
Author: cook2wj
Session Name: CGEMS Human WCSK test
Genome Assembly: hg38
Creation Date: 2016-07-21
Views: 1824
|
1469088000 |
1824 |
 |
Description:
Author: bravonja
Session Name: CGEMS Human JBSP test
Genome Assembly: hg38
Creation Date: 2016-07-21
Views: 1894
|
1469088000 |
1894 |
 |
Description:
Author: stjacqrm
Session Name: CGEMS Yeast RMSJ test
Genome Assembly: sacCer3
Creation Date: 2016-07-21
Views: 1906
|
1469088000 |
1906 |
 |
Description:
Author: s.jurg96
Session Name: CGEMS worm SJ EA CG test
Genome Assembly: ce11
Creation Date: 2016-07-21
Views: 1835
|
1469088000 |
1835 |
 |
Description:
Author: youngbh
Session Name: CGEMS Mouse BHY Custom Tracks Chr6
Genome Assembly: mm9
Creation Date: 2016-07-21
Views: 1915
|
1469088000 |
1915 |
 |
Description:
Author: youngbh
Session Name: CGEMS Mouse BHY test
Genome Assembly: mm9
Creation Date: 2016-07-21
Views: 2104
|
1469088000 |
2104 |
 |
Description:
Author: youngbh
Session Name: CGEMS Human BHY test
Genome Assembly: hg38
Creation Date: 2016-07-21
Views: 1979
|
1469088000 |
1979 |
 |
Description:
Author: youngbh
Session Name: CGEMS Worm BHY test
Genome Assembly: ce11
Creation Date: 2016-07-21
Views: 1865
|
1469088000 |
1865 |
 |
Description:
Author: dtautz
Session Name: wildmouse
Genome Assembly: mm10
Creation Date: 2016-07-26
Views: 2878
|
1469520000 |
2878 |
 |
Description:
Author: youngbh
Session Name: CGEMS Yeast BHY test
Genome Assembly: sacCer3
Creation Date: 2016-07-20
Views: 1906
|
1469001600 |
1906 |
 |
Description:
Author: arem
Session Name: colon methylation
Genome Assembly: hg19
Creation Date: 2019-03-06
Views: 1590
|
1551859200 |
1590 |
Screenshot not available
Click Here to view |
Description:
Author: Courtney Stout
Session Name: CGems Yeast AH test
Genome Assembly: sacCer3
Creation Date: 2016-07-19
Views: 1925
|
1468915200 |
1925 |
 |
Description:
Author: MWVermunt
Session Name: H3K27ac Human Brain, Vermunt et al (2014) Cell Reports
Genome Assembly: hg19
Creation Date: 2016-07-13
Views: 2182
|
1468396800 |
2182 |
 |
Description:
Author: pliu
Session Name: PRAM_master_set
Genome Assembly: hg38
Creation Date: 2019-01-04
Views: 1885
|
1546588800 |
1885 |
 |
Description:
Author: sajjeev
Session Name: comic sj15_22
Genome Assembly: hg19
Creation Date: 2019-01-04
Views: 1653
|
1546588800 |
1653 |
 |
Description:
Author: lijin
Session Name: session7 97L 3 chip
Genome Assembly: hg19
Creation Date: 2016-06-20
Views: 2151
|
1466409600 |
2151 |
 |
Description: This session displays the locations of significant SNPs from the GWAS for breast cancer DFS with P < 1 x 10-5 and eQTL-SNPs from GTEx portal with P < 1 × 10–10.
Author: Wen-Cheng
Session Name: rs1024176
Genome Assembly: hg19
Creation Date: 2018-05-25
Views: 1616
|
1527235200 |
1616 |
 |
Description: This session demonstrates the GTEx track on the human assembly hg19. The GTEx track shows data from the NIH Genotype-Tissue Expression project and displays expression data for each gene, based on GENCODE gene models, from 51 tissues collected from 570 individual. This session also demonstrates the gene-only mode of the multi-region feature, which removes intergenic regions from the display.
Author: mspeir
Session Name: hg19_gtexAnnouncement
Genome Assembly: hg19
Creation Date: 2016-06-17
Views: 2024
|
1466150400 |
2024 |
 |
Description: This session is demonstrating the exon-only mode of the multi-region feature. The exon-only mode uses the GENCODE v22 track to slice up the normal display and remove both intronic and intergenic regions from the display. Only those regions covered by exons, both coding and noncoding, are left in the display. For more on the multi-region display see the user guide: http://genome.ucsc.edu/goldenPath/help/multiRegionHelp.html.
Author: mspeir
Session Name: hg38_exonOnlyExample
Genome Assembly: hg38
Creation Date: 2016-06-17
Views: 1908
|
1466150400 |
1908 |
 |
Description:
Author: mspeir
Session Name: hg19_CAGrepeat
Genome Assembly: hg19
Creation Date: 2016-06-17
Views: 1986
|
1466150400 |
1986 |
 |
Description: Public session showcasing the GTEx gene expression track and GTEx RNA-seq signal hub for the <a target=_blank href='http://genome.ucsc.edu/blog/gtex-resources-in-the-browser/'>GTEx Resources in the Browser</a> GB blog article. The session shows a genomic region with 5 genes with different patterns of tissue-specific expression.
Author: kate
Session Name: GTEx demo for blog (long, with error)
Genome Assembly: hg19
Creation Date: 2016-06-09
Views: 4324
|
1465459200 |
4324 |
 |
Description:
Author: ruhollah
Session Name: hg19_681_chr15_nmtf13
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1681
|
1554105600 |
1681 |
 |
Description: Session highlights a mouse-specific repeat on chromosome 12. You can see the gap in the 60-way vertebrate alignment surrounding the repeat, which is highlighted in light blue. The repeat is classified as a LINE and is part of the L1 family of repeats. Additionally, on the far right-hand side of the display, you can see the retrotransposed Bf3 gene.
Author: mspeir
Session Name: mm10_MouseSpecificRepeat_plus_RetroposedGene
Genome Assembly: mm10
Creation Date: 2016-05-24
Views: 1802
|
1464076800 |
1802 |
 |
Description: Session displays the promoter region of DARC on human assembly hg19 including UCSC Genes, dbSNP 146 (Common subset) and the Publications track showing sequences and SNPs text-mined from PubMed Central and Elsevier. Adapted from Figure 1 in Meyer, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013 Jan;41(Database issue):D64-9: http://nar.oxfordjournals.org/content/41/D1/D64.long.
Author: mspeir
Session Name: hg19_NAR_2013_Fig1
Genome Assembly: hg19
Creation Date: 2016-05-24
Views: 1981
|
1464076800 |
1981 |
 |
Description: Session displays the dyskeratosis congenita 1 (DKC1) gene using the exon-only multi-region mode in the human assembly hg38. Alongside the gene structure, tracks displaying dbSNP "flagged" variants, OMIM clinical variants, ENCODE transcription levels from 9 cell lines, a 100-way vertebrate alignment (only a subset of species displayed), and conservation scores calculated using phyloP across this 100-way alignment.
Author: mspeir
Session Name: hg38_DKC1_SNPs_Cons_Transcription
Genome Assembly: hg38
Creation Date: 2016-05-24
Views: 1922
|
1464076800 |
1922 |
 |
Description: Session displays the promoter region and transcription start of IRF1 on human assembly hg19 showing UCSC Genes, 1000 Genomes Phase 3 Integrated Variant Calls in the haplotype sorting VCF display mode, histone mark H3K27Ac binding in overlays of 7 ENCODE cell lines and PhyloP conservation scores from alignments of 100 vertebrates. Adapted from Figure 2 in Meyer, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013 Jan;41(Database issue):D64-9: http://nar.oxfordjournals.org/content/41/D1/D64.long.
Author: mspeir
Session Name: hg19_NAR_2013_Fig2
Genome Assembly: hg19
Creation Date: 2016-05-24
Views: 2045
|
1464076800 |
2045 |
 |
Description:
Author: as00419
Session Name: hg38
Genome Assembly: hg38
Creation Date: 2019-05-09
Views: 1596
|
1557388800 |
1596 |
 |
Description:
Author: lijin
Session Name: session2 four types
Genome Assembly: hg19
Creation Date: 2016-04-17
Views: 2593
|
1460880000 |
2593 |
 |
Description:
Author: lijin
Session Name: session5 whole genome methy
Genome Assembly: hg19
Creation Date: 2016-04-29
Views: 2025
|
1461916800 |
2025 |
 |
Description:
Author: leipinji
Session Name: hg19-HCT116-liyiming
Genome Assembly: hg19
Creation Date: 2016-04-12
Views: 1801
|
1460448000 |
1801 |
 |
Description:
Author: Julia H-Z
Session Name: Cancer Cell Tracks
Genome Assembly: hg19
Creation Date: 2019-02-19
Views: 1641
|
1550563200 |
1641 |
 |
Description:
Author: Julia H-Z
Session Name: CC Tracks 2
Genome Assembly: hg19
Creation Date: 2019-02-20
Views: 1579
|
1550649600 |
1579 |
 |
Description:
Author: jdf228
Session Name: PGC-1 alpha ChIP-seq C2C12
Genome Assembly: mm9
Creation Date: 2017-03-20
Views: 2050
|
1489996800 |
2050 |
 |
Description:
Author: gunasekaran
Session Name: Gunasekaran_Dhandapani_UCSC_mm10
Genome Assembly: mm10
Creation Date: 2019-02-07
Views: 1533
|
1549526400 |
1533 |
 |
Description:
Author: ascendgene
Session Name: 60genes panel chr1
Genome Assembly: hg19
Creation Date: 2018-07-25
Views: 1936
|
1532505600 |
1936 |
 |
Description:
Author: peijinlim
Session Name: mm9 nrf2chipseq tracks
Genome Assembly: mm9
Creation Date: 2016-10-19
Views: 1924
|
1476864000 |
1924 |
 |
Description:
Author: SCRB20
Session Name: SCRB20_GOOD
Genome Assembly: hg19
Creation Date: 2016-03-21
Views: 1795
|
1458547200 |
1795 |
 |
Description:
Author: msantos.13
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2019-02-06
Views: 1557
|
1549440000 |
1557 |
 |
Description:
Author: janzop
Session Name: mm9_HBx-RFP
Genome Assembly: mm9
Creation Date: 2016-09-27
Views: 1765
|
1474963200 |
1765 |
 |
Description:
Author: lijin
Session Name: session1 Liver 97L exp meth
Genome Assembly: hg19
Creation Date: 2016-04-17
Views: 1985
|
1460880000 |
1985 |
 |
Description:
Author: labrozam
Session Name: mm9
Genome Assembly: mm9
Creation Date: 2016-10-02
Views: 2379
|
1475395200 |
2379 |
 |
Description:
Author: wenzelmk
Session Name: BIO 481 hg38 CHR2 Chimp Assignement
Genome Assembly: hg38
Creation Date: 2017-10-10
Views: 1669
|
1507622400 |
1669 |
 |
Description:
Author: kianoush
Session Name: Panels
Genome Assembly: hg19
Creation Date: 2019-03-06
Views: 1374
|
1551859200 |
1374 |
 |
Description:
Author: arem
Session Name: hg19_chr10_Human
Genome Assembly: hg19
Creation Date: 2019-03-08
Views: 1556
|
1552032000 |
1556 |
 |
Description:
Author: ephong0305
Session Name: KSG GWAS IA chr1_22 v1
Genome Assembly: hg19
Creation Date: 2021-07-30
Views: 1085
|
1627632000 |
1085 |
 |
Description:
Author: dunlapea
Session Name: Dunlap, BIO480 Uba1 example
Genome Assembly: hg19
Creation Date: 2016-09-04
Views: 1948
|
1472976000 |
1948 |
 |
Description:
Author: cocopow
Session Name: hg19epigenetics_WASHU_VizHub
Genome Assembly: hg19
Creation Date: 2019-02-07
Views: 1635
|
1549526400 |
1635 |
 |
Description:
Author: harri5al
Session Name: CGEMS yeast (AH) test
Genome Assembly: sacCer3
Creation Date: 2018-01-08
Views: 1912
|
1515398400 |
1912 |
 |
Description:
Author: anneabraham1
Session Name: Spring18 Yeast test Abraham
Genome Assembly: sacCer3
Creation Date: 2018-01-08
Views: 1723
|
1515398400 |
1723 |
 |
Description:
Author: nedaronaghi
Session Name: hg19-all_weeks
Genome Assembly: hg19
Creation Date: 2017-03-07
Views: 1920
|
1488873600 |
1920 |
 |
Description:
Author: Natedawg34
Session Name: fall17 bio630 human NM
Genome Assembly: hg38
Creation Date: 2017-09-13
Views: 1723
|
1505289600 |
1723 |
 |
Description:
Author: Natedawg34
Session Name: Fall 2017 Bio 630 NM
Genome Assembly: hg38
Creation Date: 2017-09-13
Views: 1663
|
1505289600 |
1663 |
 |
Description:
Author: stjacqrm
Session Name: Fall17 Bio630 Yeast RMSJ test
Genome Assembly: sacCer3
Creation Date: 2017-09-13
Views: 1698
|
1505289600 |
1698 |
 |
Description:
Author: alexc
Session Name: MitoGenome Analysis
Genome Assembly: hg38
Creation Date: 2016-05-17
Views: 1781
|
1463472000 |
1781 |
 |
Description:
Author: Natedawg34
Session Name: Miller, Fall17, Uba1
Genome Assembly: mm10
Creation Date: 2017-09-12
Views: 1748
|
1505203200 |
1748 |
 |
Description:
Author: alexc
Session Name: ARUP_cyto
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 2223
|
1543219200 |
2223 |
 |
Description: chr2
Author: diazacex
Session Name: Chimp
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 208
|
1726560000 |
208 |
 |
Description:
Author: leipinji
Session Name: hg19-HCT16-H3K27ac-P53-EP300
Genome Assembly: hg19
Creation Date: 2015-10-22
Views: 1888
|
1445500800 |
1888 |
 |
Description:
Author: hfw824
Session Name: Dou_LADs IgHV
Genome Assembly: hg19
Creation Date: 2015-10-20
Views: 2072
|
1445328000 |
2072 |
 |
Description:
Author: delphine.rolando10
Session Name: hg19_HI-LNCs_UCSCsession
Genome Assembly: hg19
Creation Date: 2016-11-01
Views: 1969
|
1477987200 |
1969 |
Screenshot not available
Click Here to view |
Description:
Author: wentao li
Session Name: 2CPD and 64 NT2
Genome Assembly: hg19
Creation Date: 2015-10-06
Views: 1752
|
1444118400 |
1752 |
 |
Description:
Author: leipinji
Session Name: hg19-hct116-LYM-RNASeq
Genome Assembly: hg19
Creation Date: 2017-10-05
Views: 1926
|
1507190400 |
1926 |
 |
Description:
Author: lianeslaughter
Session Name: mm9-H4K16acRestored
Genome Assembly: mm9
Creation Date: 2017-09-26
Views: 2212
|
1506412800 |
2212 |
 |
Description:
Author: robert-cornell@uiowa.edu
Session Name: evo mels hg19 TFAP2A in hNCC
Genome Assembly: hg19
Creation Date: 2014-05-29
Views: 1466
|
1401350400 |
1466 |
 |
Description:
Author: GenePeeks
Session Name: .2017.10 Haplotype & HGDP
Genome Assembly: hg19
Creation Date: 2017-10-06
Views: 2057
|
1507276800 |
2057 |
Screenshot not available
Click Here to view |
Description:
Author: jcasper
Session Name: bedtoolsvssamtools
Genome Assembly: hg18
Creation Date: 2014-05-27
Views: 1984
|
1401177600 |
1984 |
 |
Description:
Author: jverploegen
Session Name: GHE sequencing steroids
Genome Assembly: hg19
Creation Date: 2017-10-10
Views: 1877
|
1507622400 |
1877 |
 |
Description:
Author: hs523
Session Name: mm10-ZFP57-SentToAnastasia-merged
Genome Assembly: mm10
Creation Date: 2016-01-13
Views: 2028
|
1452672000 |
2028 |
 |
Description:
Author: GenePeeks
Session Name: .2017 Zoomed-in View 1
Genome Assembly: hg19
Creation Date: 2017-10-09
Views: 1677
|
1507536000 |
1677 |
 |
Description:
Author: taly
Session Name: EnhancerPrediction
Genome Assembly: dm3
Creation Date: 2017-10-10
Views: 1842
|
1507622400 |
1842 |
 |
Description:
Author: rhead
Session Name: 450k.EPIC.beadchip
Genome Assembly: hg19
Creation Date: 2016-11-06
Views: 2046
|
1478419200 |
2046 |
 |
Description: This mailing question allows one to see how you can color the current DNA with data from tracks such as SNP tracks. Load the session, then type "v d" or go to the View menu and then select DNA.<br>
Once you are on the "Get DNA for" page do not click "get DNA" rather click the <b>'extended case/color options'</b> button and get Extended DNA Case/Color Output with DNA colored per the SNP tracks (150 in this case) or choose other tracks. <b>Note</b> that the All SNPs are Italic and Blue and the Common SNPs are Bold and Red and when they are both the colors and style combine.
Author: brianlee
Session Name: hg19.MLQ_20306
Genome Assembly: hg19
Creation Date: 2017-10-11
Views: 1832
|
1507708800 |
1832 |
 |
Description:
Author: robert-cornell@uiowa.edu
Session Name: eliz leslie hg19
Genome Assembly: hg19
Creation Date: 2014-01-17
Views: 1481
|
1389945600 |
1481 |
 |
Description: This is a session with some highlights.
Author: brianlee
Session Name: moreHighlights
Genome Assembly: hg38
Creation Date: 2019-05-07
Views: 1521
|
1557216000 |
1521 |
 |
Description:
Author: josoga2
Session Name: chromHmm
Genome Assembly: hg19
Creation Date: 2020-06-09
Views: 1439
|
1591689600 |
1439 |
 |
Description: nar
Author: as00419
Session Name: example
Genome Assembly: hg38
Creation Date: 2019-05-09
Views: 1576
|
1557388800 |
1576 |
 |
Description:
Author: ccpowell
Session Name: 50120_solLyc1
Genome Assembly: hub_2224691_solLyc1
Creation Date: 2020-05-07
Views: 1365
|
1588838400 |
1365 |
 |
Description:
Author: bashamal
Session Name: hg38 chr2 comparison 10/10 in class
Genome Assembly: hg38
Creation Date: 2017-10-10
Views: 1764
|
1507622400 |
1764 |
 |
Description:
Author: estercastillo
Session Name: VanesaGarcia_ClinicoSanCarlos_hotspots_131686
Genome Assembly: hg19
Creation Date: 2017-10-19
Views: 1766
|
1508400000 |
1766 |
 |
Description:
Author: cocopow
Session Name: hg18_44way_bed
Genome Assembly: hg18
Creation Date: 2019-07-24
Views: 1440
|
1563955200 |
1440 |
 |
Description:
Author: wenzelmk
Session Name: BIO481 Mouse (Wenzel & Zeher) test
Genome Assembly: mm9
Creation Date: 2017-10-05
Views: 1596
|
1507190400 |
1596 |
 |
Description:
Author: macdonem
Session Name: Bio481 Mouse MacDonald Basham Test
Genome Assembly: mm9
Creation Date: 2017-10-05
Views: 1686
|
1507190400 |
1686 |
 |
Description:
Author: herdaj
Session Name: Bio481 Human Herd Test
Genome Assembly: hg38
Creation Date: 2017-10-05
Views: 1853
|
1507190400 |
1853 |
 |
Description:
Author: labrozam
Session Name: Bio481 Human Labrozzi-Grant test
Genome Assembly: hg38
Creation Date: 2017-10-05
Views: 1703
|
1507190400 |
1703 |
 |
Description:
Author: XiangChen
Session Name: Bio481 Yeast Chen Test
Genome Assembly: sacCer3
Creation Date: 2017-10-05
Views: 1708
|
1507190400 |
1708 |
 |
Description: This session shows the Integrative and Discriminative Epigenome Annotation System (IDEAS) Public Hub, which has data for inferred chromatin states in 127 cell types. A color key for each state is shown near a heatmap on the Track Description page, also found in the related journal <a href="https://academic.oup.com/view-large/figure/97265435/gkx659fig1.jpg/Accurate%20and%20reproducible%20functional%20maps%20in%20127%20human%20cell%20types%20via%202D%20genome%20segmentation">figure 1</a>. This session has some segmentation examples by IDEAS and ChromHMM in HSC and B-cell cell types (9 total) near genes CIITA and CLEC16A. Find more reference information on the Track Description page or at this <a href="https://doi.org/10.1093/nar/gkx659">journal article</a>. The IDEAS algorithm is available on GitHub at <a href="https://github.com/yuzhang123/IDEAS">https://github.com/yuzhang123/IDEAS</a>.
Author: brianlee
Session Name: hg19.IDEAS.hub
Genome Assembly: hg19
Creation Date: 2017-10-05
Views: 2199
|
1507190400 |
2199 |
 |
Description:
Author: estercastillo
Session Name: VanesaGarcia_ClinicoSanCarlos_Pancreas_131774
Genome Assembly: hg19
Creation Date: 2017-10-22
Views: 1756
|
1508659200 |
1756 |
 |
Description:
Author: hujian5241
Session Name: fly_dh1_dm3_bw
Genome Assembly: dm3
Creation Date: 2017-11-13
Views: 1710
|
1510560000 |
1710 |
 |
Description:
Author: simonwang612
Session Name: Methylation
Genome Assembly: mm10
Creation Date: 2020-07-25
Views: 1853
|
1595664000 |
1853 |
 |
Description:
Author: anneabraham1
Session Name: Abraham, Spring18, Uba1 example
Genome Assembly: mm9
Creation Date: 2018-01-13
Views: 1899
|
1515830400 |
1899 |
 |
Description:
Author: robert-cornell@uiowa.edu
Session Name: Ruchi tracks hg19
Genome Assembly: hg19
Creation Date: 2014-01-27
Views: 1481
|
1390809600 |
1481 |
 |
Description:
Author: twx123
Session Name: PTC-mm9-newest
Genome Assembly: mm9
Creation Date: 2015-06-10
Views: 1593
|
1433923200 |
1593 |
 |
Description:
Author: cocopow
Session Name: hg38_chr17_KCNJ18
Genome Assembly: hg38
Creation Date: 2019-03-07
Views: 1743
|
1551945600 |
1743 |
 |
Description:
Author: robert-cornell@uiowa.edu
Session Name: mm9 visel enhancers
Genome Assembly: mm9
Creation Date: 2014-01-11
Views: 1556
|
1389427200 |
1556 |
 |
Description:
Author: Shirley
Session Name: WT-RNA-ribo-seq
Genome Assembly: mm10
Creation Date: 2018-11-20
Views: 1441
|
1542700800 |
1441 |
 |
Description:
Author: QAtester
Session Name: 111hg38
Genome Assembly: hg38
Creation Date: 2018-12-21
Views: 1951
|
1545379200 |
1951 |
 |
Description:
Author: Boonede
Session Name: MAPT_DougBoone
Genome Assembly: hg38
Creation Date: 2019-03-11
Views: 1569
|
1552291200 |
1569 |
 |
Description:
Author: Guillermo6
Session Name: hg19_ashg2014_SNPs
Genome Assembly: hg19
Creation Date: 2017-11-01
Views: 1883
|
1509523200 |
1883 |
 |
Description:
Author: ferhatay
Session Name: hg19
Genome Assembly: hg19
Creation Date: 2016-08-25
Views: 1867
|
1472112000 |
1867 |
 |
Description:
Author: kkarri
Session Name: rn6_transcriptome
Genome Assembly: rn6
Creation Date: 2017-10-27
Views: 1843
|
1509091200 |
1843 |
 |
Description:
Author: wangt5
Session Name: HBEC_VitD
Genome Assembly: hg19
Creation Date: 2017-11-15
Views: 1695
|
1510732800 |
1695 |
 |
Description:
Author: bashamal
Session Name: Basham, Fall17, Uba1 example
Genome Assembly: mm9
Creation Date: 2017-10-02
Views: 1751
|
1506931200 |
1751 |
 |
Description:
Author: cocapo
Session Name: ponAbe3_BTN1A1
Genome Assembly: ponAbe3
Creation Date: 2018-11-15
Views: 1841
|
1542268800 |
1841 |
 |
Description:
Author: lbenner
Session Name: Sxl
Genome Assembly: dm6
Creation Date: 2016-10-22
Views: 2113
|
1477123200 |
2113 |
 |
Description:
Author: macdonem
Session Name: Old AMD SNPs
Genome Assembly: hg38
Creation Date: 2017-10-31
Views: 1704
|
1509436800 |
1704 |
 |
Description:
Author: macdonem
Session Name: New AMD SNPs
Genome Assembly: hg38
Creation Date: 2017-10-31
Views: 1722
|
1509436800 |
1722 |
 |
Description:
Author: falbert
Session Name: FP_sacCer3
Genome Assembly: sacCer3
Creation Date: 2013-08-07
Views: 1729
|
1375862400 |
1729 |
 |
Description:
Author: ruhollah
Session Name: hg19_94Chr12_28
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1724
|
1543219200 |
1724 |
 |
Description: EEPD1 ChIP-seq on chromosome 1 hg38
Author: abacolla
Session Name: eepd1_chr1
Genome Assembly: hg38
Creation Date: 2023-05-30
Views: 537
|
1685433600 |
537 |
 |
Description:
Author: cocopow
Session Name: hg38_ RepeatMasker
Genome Assembly: hg38
Creation Date: 2019-01-24
Views: 1669
|
1548316800 |
1669 |
 |
Description:
Author: kkaczor
Session Name: cirm_ucla
Genome Assembly: hg38
Creation Date: 2020-11-12
Views: 1352
|
1605168000 |
1352 |
 |
Description:
Author: cocopow
Session Name: hg38_tf _pc
Genome Assembly: hg38
Creation Date: 2019-06-11
Views: 1481
|
1560240000 |
1481 |
 |
Description:
Author: cocopow
Session Name: hg38_tf_pc2
Genome Assembly: hg38
Creation Date: 2019-06-11
Views: 1429
|
1560240000 |
1429 |
 |
Description:
Author: cocopow
Session Name: hg19_myc+ctcf
Genome Assembly: hg19
Creation Date: 2019-06-11
Views: 1605
|
1560240000 |
1605 |
 |
Description:
Author: cocopow
Session Name: hg38_myc+ctcf
Genome Assembly: hg38
Creation Date: 2019-06-11
Views: 1599
|
1560240000 |
1599 |
 |
Description:
Author: dtautz
Session Name: wildmouse-introgression
Genome Assembly: mm10
Creation Date: 2017-11-04
Views: 1812
|
1509782400 |
1812 |
 |
Description:
Author: lijin
Session Name: session4 plus h3k27ac density andpeaks
Genome Assembly: hg19
Creation Date: 2016-04-28
Views: 1897
|
1461830400 |
1897 |
 |
Description:
Author: lijin
Session Name: session3 plus h3k27ac
Genome Assembly: hg19
Creation Date: 2016-04-28
Views: 1970
|
1461830400 |
1970 |
 |
Description:
Author: XiangChen
Session Name: Bio481 Yeast Chen Custom Track
Genome Assembly: sacCer3
Creation Date: 2017-10-17
Views: 1798
|
1508227200 |
1798 |
 |
Description:
Author: XiangChen
Session Name: Bio481 Chen Gzmm CBRs in Mice Photoreceptors
Genome Assembly: mm9
Creation Date: 2017-11-17
Views: 1656
|
1510905600 |
1656 |
 |
Description: This session illustrates how a user was having a hard time aligning the cDNA sequence with their Predicted Protein. They were manually using DNA sequence to make a triplet number to figure out codon numbering. The session shows how when zoomed in on the base level, if you right click the top Base Position track and set it to display in "full" you will see predicted proteins for every set of codons across the genome. This particular session at the start of the SIRT1 gene is also displaying the conservation tracks with similar coding information for several species. This information is also available in the table browser, where you can select output as "sequence" for a genomic region when you select a gene's track table as the source.
Author: brianlee
Session Name: exampleProteinGene
Genome Assembly: hg19
Creation Date: 2013-08-23
Views: 439
|
1377244800 |
439 |
 |
Description: HBB on hg38 with single cell Heart and Merged data
Author: ashg2023
Session Name: hg38_HBB_singleCell1
Genome Assembly: hg38
Creation Date: 2023-08-03
Views: 433
|
1691049600 |
433 |
 |
Description: Hemoglobin beta 3 isoforms displayed, snp155 Common turned on.
Author: ashg2023
Session Name: hg38_HBB_snp155
Genome Assembly: hg38
Creation Date: 2023-08-03
Views: 444
|
1691049600 |
444 |
 |
Description:
Author: jwjiao
Session Name: E13-forebrain-H3K36me3-ChIP
Genome Assembly: mm10
Creation Date: 2017-11-06
Views: 1927
|
1509955200 |
1927 |
 |
Description:
Author: ruhollah
Session Name: hg19_45Chr3_nmt10
Genome Assembly: hg19
Creation Date: 2018-11-27
Views: 1563
|
1543305600 |
1563 |
 |
Description:
Author: ruhollah
Session Name: hg19_56Chr19_nmt10
Genome Assembly: hg19
Creation Date: 2018-11-27
Views: 1542
|
1543305600 |
1542 |
 |
Description:
Author: isilver1
Session Name: PABPC1_CLIP_Public
Genome Assembly: mm10
Creation Date: 2015-07-01
Views: 2576
|
1435737600 |
2576 |
 |
Description:
Author: xiaotiao
Session Name: 4CProject
Genome Assembly: hg38
Creation Date: 2019-03-15
Views: 1478
|
1552636800 |
1478 |
 |
Description:
Author: zhangjing12
Session Name: 201901-ATAC seq
Genome Assembly: hg38
Creation Date: 2019-03-15
Views: 1566
|
1552636800 |
1566 |
 |
Description: human chr.2 synteny to other primates
Author: marte2je
Session Name: hg38 to other primates - chr2
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 202
|
1726560000 |
202 |
 |
Description: 9/17/24
Author: aehodges
Session Name: hg38 chromosome 2/Chimp
Genome Assembly: hg38
Creation Date: 2024-09-17
Views: 196
|
1726560000 |
196 |
 |
Description:
Author: hujian5241
Session Name: fly_dh1_dm3
Genome Assembly: dm3
Creation Date: 2017-11-12
Views: 1676
|
1510473600 |
1676 |
 |
Description:
Author: salima
Session Name: e32_tailseq_5p_3p
Genome Assembly: ce10
Creation Date: 2015-07-03
Views: 2212
|
1435910400 |
2212 |
 |
Description: This session was used to answer a mailing list question about mm9.retina data. You can see how the signal data was processed to create finalized peaks.
Author: brianlee
Session Name: mm9.retina
Genome Assembly: mm9
Creation Date: 2017-11-14
Views: 1718
|
1510646400 |
1718 |
 |
Description:
Author: keitermd
Session Name: Keiter, Spring 2017, Uba1 example
Genome Assembly: mm9
Creation Date: 2017-01-15
Views: 1772
|
1484467200 |
1772 |
 |
Description:
Author: aurbanow
Session Name: srivatsan
Genome Assembly: hg38
Creation Date: 2020-12-28
Views: 1502
|
1609142400 |
1502 |
 |
Description:
Author: nicklewis
Session Name: photoreceptor sesh
Genome Assembly: mm9
Creation Date: 2017-11-16
Views: 1790
|
1510819200 |
1790 |
 |
Description:
Author: nicklewis
Session Name: photorecptor session
Genome Assembly: mm9
Creation Date: 2017-11-16
Views: 1720
|
1510819200 |
1720 |
 |
Description:
Author: nrhong
Session Name: hg38_KIAA1217
Genome Assembly: hg38
Creation Date: 2018-01-25
Views: 1725
|
1516867200 |
1725 |
 |
Description:
Author: labrozam
Session Name: wt/Nrl-
Genome Assembly: mm9
Creation Date: 2017-11-17
Views: 1720
|
1510905600 |
1720 |
 |
Description:
Author: german.n
Session Name: zebrafish_Iso-Seq
Genome Assembly: danRer10
Creation Date: 2018-03-16
Views: 1736
|
1521187200 |
1736 |
 |
Description:
Author: ruhollah
Session Name: hg19_1220_chr3_nmtf13
Genome Assembly: hg19
Creation Date: 2019-04-01
Views: 1706
|
1554105600 |
1706 |
 |
Description:
Author: Yog77
Session Name: chrMT_rCRS_NC012920
Genome Assembly: hg19
Creation Date: 2020-05-20
Views: 1353
|
1589961600 |
1353 |
 |
Description:
Author: timmnat
Session Name: Figure4-Differences_in_polymorphism_frequency_and_copy number.hub_1106737_Amex_PQ.v4
Genome Assembly: hub_1106737_Amex_PQ.v4
Creation Date: 2020-09-13
Views: 1672
|
1599984000 |
1672 |
 |
Description:
Author: robert-cornell@uiowa.edu
Session Name: mm9 Loftus/Pavan Tfap2a
Genome Assembly: mm9
Creation Date: 2014-06-23
Views: 1464
|
1403510400 |
1464 |
 |
Description:
Author: robert-cornell@uiowa.edu
Session Name: tfap2a in Ncc, MITF hg18
Genome Assembly: hg18
Creation Date: 2014-09-30
Views: 1549
|
1412064000 |
1549 |
 |
Description:
Author: grant2jm
Session Name: Grant, Fall 17, allignment example
Genome Assembly: hg38
Creation Date: 2017-10-09
Views: 1694
|
1507536000 |
1694 |
 |
Description:
Author: katiegal
Session Name: Lhx3-Isl1-p53
Genome Assembly: mm9
Creation Date: 2014-10-01
Views: 1381
|
1412150400 |
1381 |
 |
Description:
Author: mattievaz8
Session Name: 062816_Mattie
Genome Assembly: mm10
Creation Date: 2016-06-28
Views: 1943
|
1467100800 |
1943 |
 |
Description:
Author: nicklewis
Session Name: Lewis Fall17 Uba1
Genome Assembly: mm9
Creation Date: 2017-10-03
Views: 1743
|
1507017600 |
1743 |
 |
Description:
Author: holtonke
Session Name: CGEMS Worm KEH test
Genome Assembly: ce11
Creation Date: 2016-07-21
Views: 1920
|
1469088000 |
1920 |
 |
Description:
Author: alyssa
Session Name: Ayala-Ps2-ACE2-session
Genome Assembly: hg19
Creation Date: 2020-05-07
Views: 1661
|
1588838400 |
1661 |
 |
Description:
Author: yierjin_2008
Session Name: MMCL.H3K27ac.SEs
Genome Assembly: hg19
Creation Date: 2017-11-10
Views: 1882
|
1510300800 |
1882 |
 |
Description:
Author: ruhollah
Session Name: hg19_72Chr14_nmt14
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 2056
|
1543219200 |
2056 |
 |
Description:
Author: ShiyiYin
Session Name: mega data with coordinations
Genome Assembly: hg19
Creation Date: 2017-02-08
Views: 1948
|
1486540800 |
1948 |
 |
Description:
Author: ShiyiYin
Session Name: intron6 ehf pcr
Genome Assembly: hg19
Creation Date: 2017-01-27
Views: 1941
|
1485504000 |
1941 |
 |
Description: This session highlights the "Seq-data on mm9 NS5 cells" public hub, where Juan Mateo et al. assayed crucial transcription factor binding activity forming the basis of the neural stem cell self-renewal regulatory network.
Author: chmalee
Session Name: mm9_NS5_Cell_Expression
Genome Assembly: mm9
Creation Date: 2017-11-28
Views: 2261
|
1511856000 |
2261 |
 |
Description:
Author: grant2jm
Session Name: Grant, Fall17, Uba1 example
Genome Assembly: mm9
Creation Date: 2017-10-02
Views: 1654
|
1506931200 |
1654 |
 |
Description:
Author: isilver1
Session Name: NAD
Genome Assembly: mm10
Creation Date: 2015-03-09
Views: 2069
|
1425888000 |
2069 |
 |
Description:
Author: ruhollah
Session Name: 1_50_outliers_hg19
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1878
|
1543219200 |
1878 |
 |
Description:
Author: zheyangshan
Session Name: hg19 GRO-seq BCL2
Genome Assembly: hg19
Creation Date: 2019-01-16
Views: 1458
|
1547625600 |
1458 |
 |
Description:
Author: mspeir
Session Name: hg19_wobble
Genome Assembly: hg19
Creation Date: 2016-06-17
Views: 2522
|
1466150400 |
2522 |
 |
Description:
Author: toni.berger@medisin.uio.no
Session Name: example1
Genome Assembly: hg19
Creation Date: 2017-11-13
Views: 2036
|
1510560000 |
2036 |
 |
Description:
Author: puertoRico2019
Session Name: DQP
Genome Assembly: hg19
Creation Date: 2019-04-12
Views: 1420
|
1555056000 |
1420 |
 |
Description:
Author: mziegler
Session Name: encode_Accessibility+TAD
Genome Assembly: hg19
Creation Date: 2018-09-11
Views: 1594
|
1536652800 |
1594 |
 |
Description:
Author: cath
Session Name: v338test
Genome Assembly: hg38
Creation Date: 2016-09-13
Views: 1860
|
1473753600 |
1860 |
 |
Description:
Author: josoga2
Session Name: joseph_omix_hw
Genome Assembly: mm10
Creation Date: 2020-05-19
Views: 1339
|
1589875200 |
1339 |
 |
Description:
Author: macdonem
Session Name: Bio481 Mouse MacDonald Basham Custom Track
Genome Assembly: mm9
Creation Date: 2017-10-17
Views: 1681
|
1508227200 |
1681 |
 |
Description:
Author: nicklewis
Session Name: CGEMS Yeast Test - Custom Track
Genome Assembly: sacCer3
Creation Date: 2017-10-17
Views: 1704
|
1508227200 |
1704 |
 |
Description:
Author: herdaj
Session Name: Bio481 Human Custom Herd
Genome Assembly: hg38
Creation Date: 2017-10-17
Views: 1968
|
1508227200 |
1968 |
 |
Description:
Author: erick.gabriel
Session Name: MYBPC1 alleles
Genome Assembly: hg19
Creation Date: 2018-09-07
Views: 1639
|
1536307200 |
1639 |
 |
Description: This example session shows some selected cell lines displayed with DNaseI tracks available for the mm9 mouse assembly from the ENCODE project.
You can click the grey bars at the far left to go to the track descriptions, and select even more cell types.
This session is the result of a user requesting where to find known areas/regions of chromatin in the mouse genome, especially condensed regions of DNA. By entering the user's coordinates of interest the researcher could investigate data indicating hypersensitive DNase regions.
Author: brianlee
Session Name: DNase in mm9 Example
Genome Assembly: mm9
Creation Date: 2013-07-30
Views: 397
|
1375171200 |
397 |
 |
Description:
Author: ruhollah
Session Name: hg19_209Chr7_nmt4
Genome Assembly: hg19
Creation Date: 2019-01-14
Views: 1564
|
1547452800 |
1564 |
 |
Description:
Author: marzoldr
Session Name: Marzolf, Spring18, Uba1 example
Genome Assembly: mm9
Creation Date: 2018-01-17
Views: 1639
|
1516176000 |
1639 |
 |
Description: The GeneHancer track relates enhancer and promoters to their interactions with nearby genes. In this display, a highlighted GH01J209814 enhancer is associated with the gene IRF6 (Interferon Regulatory Factor 6) located about 10kb upstream from the gene and harbors regulatory non-coding variants strongly associated to Van Der Woude Syndrome 1 (VWS1), a disease involving cleft lip and cleft palate.
Author: view
Session Name: GeneHancer
Genome Assembly: hg19
Creation Date: 2019-03-29
Views: 1460
|
1553846400 |
1460 |
 |
Description:
Author: ferrerjm
Session Name: Ferrer, FALL17, yeast custom track
Genome Assembly: sacCer3
Creation Date: 2017-10-17
Views: 2269
|
1508227200 |
2269 |
 |
Description: This session displays a region of the LHX6 gene that highlights a selection of the new tracks added in the previous year for the hg38/GRCh38 human assembly. The tracks shown in this display (from top to bottom) include GENCODE Genes V22, transcription levels assayed across 9 ENCODE cell lines, DNase hypersensitive regions based on data from 95 ENCODE cell lines, genome-wide conservation scores calculated using phastCons, a multiple genome alignment created using Lastz and Multiz, and pathogenic CNVs from the ClinGen database. Adapted from Figure 1 in Speir, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016 Jan 4;44(D1):D717-25: http://nar.oxfordjournals.org/content/44/D1/D717.full
Author: mspeir
Session Name: hg38_NAR_2016_Fig1
Genome Assembly: hg38
Creation Date: 2015-08-11
Views: 2128
|
1439280000 |
2128 |
 |
Description:
Author: ruhollah
Session Name: hg19_1_chr15
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1692
|
1543219200 |
1692 |
 |
Description:
Author: ruhollah
Session Name: hg19_4_chr3
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1581
|
1543219200 |
1581 |
 |
Description:
Author: ruhollah
Session Name: hg19_3_chr14
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1550
|
1543219200 |
1550 |
 |
Description:
Author: ruhollah
Session Name: hg19_22Chr19_nmt32
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1675
|
1543219200 |
1675 |
 |
Description:
Author: ruhollah
Session Name: hg19_25Chr5_nmt29
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1523
|
1543219200 |
1523 |
 |
Description:
Author: ruhollah
Session Name: hg19_69Chr1_nmt29
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1584
|
1543219200 |
1584 |
 |
Description:
Author: ruhollah
Session Name: hg19_17Chr3_nmt26
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1581
|
1543219200 |
1581 |
 |
Description:
Author: Atma
Session Name: hervh-MYO16
Genome Assembly: hg38
Creation Date: 2019-01-22
Views: 2033
|
1548144000 |
2033 |
 |
Description: This browsing started on hg38, and then the region was lifted to hg19, and then the TFBS track turned on to show how some histone activity is related to TFBS that aren't seen on hg38 because the track isn't available there.
Author: brianlee
Session Name: hg19_MTOR
Genome Assembly: hg19
Creation Date: 2019-01-22
Views: 1579
|
1548144000 |
1579 |
 |
Description:
Author: ruhollah
Session Name: hg19_47Chr14_nmt17
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1546
|
1543219200 |
1546 |
 |
Description:
Author: ruhollah
Session Name: hg19_55Chr21_nmt19
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1568
|
1543219200 |
1568 |
 |
Description:
Author: ruhollah
Session Name: hg19_46ChrX_nmt20
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 2130
|
1543219200 |
2130 |
 |
Description:
Author: ruhollah
Session Name: hg19_31Chr22_nmt20
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1524
|
1543219200 |
1524 |
 |
Description:
Author: ruhollah
Session Name: hg19_37Chr1_nmt21
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1664
|
1543219200 |
1664 |
Screenshot not available
Click Here to view |
Description:
Author: ruhollah
Session Name: hg19_18Chr17_21
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1602
|
1543219200 |
1602 |
 |
Description:
Author: ruhollah
Session Name: hg19_16Chr5_nmt21
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1894
|
1543219200 |
1894 |
 |
Description:
Author: ruhollah
Session Name: hg19_12Chr3_nmt22
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1636
|
1543219200 |
1636 |
 |
Description:
Author: ruhollah
Session Name: hg19_95Chr16_nmt17
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1527
|
1543219200 |
1527 |
 |
Description:
Author: kinerettaler
Session Name: danRer11_plod3+-100,000
Genome Assembly: danRer11
Creation Date: 2018-12-25
Views: 1479
|
1545724800 |
1479 |
 |
Description:
Author: ruhollah
Session Name: hg19_26Chr17_nmt14
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1526
|
1543219200 |
1526 |
 |
Description:
Author: xiaotiao
Session Name: txiao
Genome Assembly: hg19
Creation Date: 2019-03-13
Views: 1592
|
1552464000 |
1592 |
 |
Description:
Author: ruhollah
Session Name: hg19_70Chr6_nmt14
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1718
|
1543219200 |
1718 |
 |
Description:
Author: ldistefano
Session Name: dm6_LDiStefanoData
Genome Assembly: dm6
Creation Date: 2019-03-12
Views: 1578
|
1552377600 |
1578 |
 |
Description:
Author: marylaws
Session Name: hg18 Mol Biol Cell 2013 Grant GD
Genome Assembly: hg18
Creation Date: 2017-11-06
Views: 1943
|
1509955200 |
1943 |
 |
Description: Acinetobacter baumannii ATCC 17978 UN
Author: Ab17978Cook
Session Name: GCF_019356215.1
Genome Assembly: hub_3861787_GCF_019356215.1
Creation Date: 2023-01-12
Views: 494
|
1673510400 |
494 |
 |
Description:
Author: ruhollah
Session Name: hg19_75Chr7_nmt16
Genome Assembly: hg19
Creation Date: 2018-11-26
Views: 1559
|
1543219200 |
1559 |
 |
Description:
Author: ruhollah
Session Name: hg19_40Chr22_nmt12
Genome Assembly: hg19
Creation Date: 2018-11-27
Views: 1587
|
1543305600 |
1587 |
 |
Description:
Author: ruhollah
Session Name: hg19_49Chr3_nmt12
Genome Assembly: hg19
Creation Date: 2018-11-27
Views: 1544
|
1543305600 |
1544 |
 |
Description:
Author: ruhollah
Session Name: hg19_58Chr1_nmt10
Genome Assembly: hg19
Creation Date: 2018-11-27
Views: 1557
|
1543305600 |
1557 |
 |
Description:
Author: ruhollah
Session Name: hg19_89Chr5_nmt10
Genome Assembly: hg19
Creation Date: 2018-11-27
Views: 1610
|
1543305600 |
1610 |
 |
Description:
Author: kazuhidew
Session Name: hg38_NAR_submission
Genome Assembly: hg38
Creation Date: 2018-11-28
Views: 1682
|
1543392000 |
1682 |